Abstract
Introduction: This study aims to confirm the association between SNP rs2046210 and breast cancer in the Vietnamese population.
Methods: A case/control study has been performed with the sample size of 300 cases and 325 controls. High Resolution Melting method is optimised for SNP genotyping. Statistic logistic regression analysis was used along with dominant and recessive analysis to analyse the association between alleles and genotypes of the selected SNP and breast cancer risk.
Results: With the selection population and the optimal HRM method, genotyping resulted in precise data for association analysis as the HWE test showed the distribution of genotypes in the population is in equilibrium (p > 0.05). The logistic regression association analysis showed SNP rs2046210 strongly associated with breast cancer risk in both allelic (p = 0.0015) and genotypic (p = 0.0064) analysis. The risk allele A, like other previous studies, strongly associated with increasing the risk of breast cancer up to 1.425 folds (OR [95%CI] = 1.425 [1.143- 1.777], p = 0.0015). However, it has shown the recessive effect in the additive analysis. Genotype AA showed stronger effect on the risk of breast cancer while the heterozygous AG genotypes showed weaker effect. Due to the small sample size, statistical power of this study is not high as expected (58.67%). But this case/control study confirms the association between SNP rs2046210 and the risk of breast cancer in the Vietnamese population.
Conclusion: SNP rs2046210 is associated with breast cancer risk in the Vietnamese population. The risk allele A plays a role in increasing the risk of breast cancer in Vietnamese women.
Introduction
Cancer is the disease that has received the most attention recently because of its complications. The mechanism of cancer is not clear. Treatment of cancer fails if the disease is found in later stages. Within cancers, breast cancer (BC) is the second most common cancer as well as the most common cancer in women worldwide. According to Globocan 2012 (WHO), BC was the third prominent cancer in Vietnam. It is expected to increase in the future and become the most prominent cancer in women Torre et al., 2015. Fighting breast cancer requires early diagnosis. According to WHO, the best protection from cancer is early detection which greatly increases the chances for successful treatment (WHO, 2016). When treated at an early stage, many patients survive at least 5 years after diagnosis (the 5-year survival rate). This rate is relatively high with patients who are diagnosed in stage 0-I: 100%; in stage II: 93%; and is very low at the late stage III: 72%; and stage IV with 22% DeSantis et al., 2013.
Breast tumorogenesis involved both extrinsic factors (e.g. environmental factors) and intrinsic factors (e.g. genetic factors). Among which about 5% to 10% of breast cancer cases are thought to be hereditary Pei et al., 2013. Besides, DNA variations appearing may be caused by intracellular exposure to endogenous and exogenous mutagens, which can cause genetic instability and carcinogenesis. Understanding about the DNA damage or variations in the cancer development is an important study which provides information for early diagnosis and genetic therapy in the future.
Single Nucleotide Polymorphism (or SNP) is a common variation with small change in the DNA sequence; however, recently its role in development of several diseases has been demonstrated. It may play a crucial role in the regulation of the gene expression, which may alter the protein level or structure leading to development of certain diseases. One SNP may contribute a bit to the development of the disease, but the haplotype of several SNPs have been known to play the main role in disease development or resistance to a certain disease Dunstan et al., 2014. GWA studies have been performed and identified several SNPs associated with cancer and particularly breast cancer Easton et al., 2007Gold et al., 2008Hunter et al., 2007Long et al., 2012Stacey et al., 2007Zheng et al., 2009. In a smaller scale, many other studies have demonstrated that some individual SNPs are very strongly associated with breast cancer. SNP rs2046210 is known as one of the very strong candidates for breast cancer association study. In 2009, Zheng et al has identified SNP rs2046210 to be highly associated with breast cancer in Asian population Zheng et al., 2009, especially in Chinese [ORs (95%CI) = 1.35(1.17-1.57), p = 4.1.10-5] (Lin et al., 2014) or [ORs (95%CI) = 1.30(1.22–1.38) and 1.64(1.50–1.80) for the AG and AA genotypes, respectively, p = 1.54×10−30]; Japanese women [ORs (95%CI) = 1.37(1.11-1.70), p = 0.05] Mizoo et al., 2013 or [ORs (95%CI) = 1.31(1.13–1.52) and 1.37(1.06–1.76), p = 2.51×10−4] Cai et al., 2011; Korean women [ORs (95%CI) = 1.31 (1.19–1.45), p = 7.91 × 10−8 for dominant model] Han et al., 2011. This SNP also has a positive association with breast cancer risk in European-ancestry American women with [ORs (95% CI) = 1.07 (0.99 – 1.16) and 1.18 (1.04 – 1.34), P for trend = 0.0069]. However, there was no association observed in African American women [ORs (95% CI) = 0.81 (0.63 – 1.06) and 0.85 (0.65 – 1.11) for the AG and AA genotypes, respectively, P for trend = 0.4027] Cai et al., 2011.
SNP rs2046210 is located in 6q25.1 locus of chromosome 6, approximately 29 kb upstream of the estrogen receptor 1 (ESR1) gene, which is responsible for expressing ERα - the cofactor of estrogen hormone. Due to its close proximity to ESR1, rs2046210 is suspected to alter ESR1 expression thereby causing uncontrolled proliferation of breast epithelial cells, which leads to breast tumor formation Dunbier et al., 2011Zhou et al., 2013. Its A-allele was associated with a population attributable risk of 18.9% and an estimated 2.1% excess familial risk of breast cancer, especially for the AG and AA genotypes Zheng et al., 2009. Nearly 60% elevated risk of breast cancer was found among women homozygous for the variant A allele in rs2046210 Cai et al., 2011. This study aimed to demonstrate the association between the SNP rs2046210 and the breast cancer in the Vietnamese population.
Methods
Subjects
300 women patients who confirmed having breast cancer and preparing for surgery were informed to participate in this study. The other group of 325 volunteers indicated as healthy persons and without breast cancer also joined this study. The blood samples were collected from all participants satisfying the sample selection criteria including matching in sex, Kinh - Vietnamese were willing to sign the consent form, which was approved by the Ethical Committee of Oncology Hospital – HCMC Vietnam under the decision number 177/HĐĐĐ-CĐT, 18th November 2014.
2 mL of whole blood samples were collected by vein puncture and put in EDTA container till DNA extraction was performed. The DNA extraction was then performed using the salting out method Hue et al., 2012 with some modifications for whole blood samples. 500 µL whole blood was used for each extraction. The DNA extracted samples were then stored at -20°C until used for PCR assay.
SNP selection
This SNP was chosen for its ubiquity in East Pacific Asian breast cancer incidence. Being one of the oldest countries in East, it is likely that Vietnam also shares the same association with rs2046210 as other Asian populations. There were many studies which demonstrated that SNP rs2046210 associated with breast cancer, thus it is possible this happens in Vietnamese. The SNP rs2046210 is selected for this study aim to confirm the association of this SNP with breast cancer, particularly in the Vietnamese population.
Genotyping method
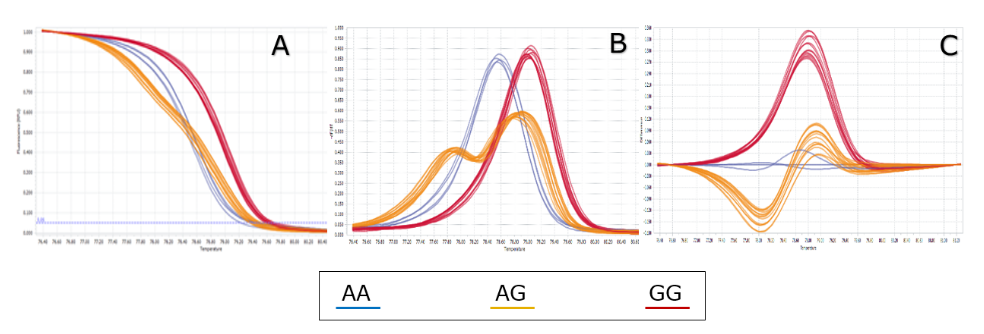
The genotyping method selected for this study is High Resolution Melting (HRM). The DNA sequence around SNP rs2046210 is obtained from the NCBI SNP database and then used for designing PCR primers using Primer3Plus. The web tool uMelt HETS (https://www.dna.utah.edu/hets/umh.php) was used for predicting the PCR products’ melting curve Wittwer et al., 2003. The best primer pair (rs2046210-F-5’-AAAGGCATGCTGGAAGAGTGTTTT-3’ and rs2046210-R-5’- GGGTGCCTCAACTGTCTTGTGA-3’) for HRM has been selected to amplify the fragment of 131 bp and give 3 distinct curves for the 3 genotypes GG, AA and GA.
The PCR was carried out in a LightCycler 96 Instrument (Roche Diagnostics, Penzberg, Germany) using Roche HRM master mix (Roche Diagnostics, Germany). Thermal cycling consisted of an initial pre-incubation at 95°C for 10 minutes, followed by 40 cycles of denaturation at 95°C for 10 s, annealing for 10 s at 64°C (Ta) and elongation at 72°C for 10 s. Finally, heteroduplexes were generated by adding a step at 95°C for 1 minute and cooling the reaction to 40°C (Ramp rate of 2.2°C/s). The best conditions for HRM analysis have been optimised with the control samples (data not shown).
Association analysis
Statistical analysis was conducted STATA ver12. The Hardy–Weinberg equilibrium, the logistic regression analysis was performed. Associations among genotypes and breast cancer risk were then estimated by computing odds ratios (ORs) and 95% confidence intervals (CIs) from the logistic regression analysis. The dominant, recessive and additive analysis was performed using website https://ihg.gsf.de/cgi-bin/hw/hwa1.pl to double check and clarify the role of allele in dominant or recessive affect. All statistical tests had a P-value of < 0.05 was considered statistically significant. Sample size and statistical power of the case-control study was computed by a genetic power calculator (http:// sampsize.sourceforge.net/iface/s3.html#ccp). Genetic Power Calculator developed by Purcell et al. (http://pngu.mgh.harvard.edu/~purcell/gpc/).
Results
Genotyping
Using the optimal HRM condition as mentioned in the method above, totally 625 samples were successfully genotyped, including 325 controls and 300 cases. Three different genotypes were identified due to different curve shapes in three analysis channels: melting curve, melting peak and different plot as described in Figure 1 . The genotyping result of 625 samples is very clear, and there is no confusion (compared to other SNPs which may appear in some confused samples – data not shown). Three different genotypes distinguished clearly along with three genotype controls ( Figure 1 ).
Alleles’ frequency and HWE
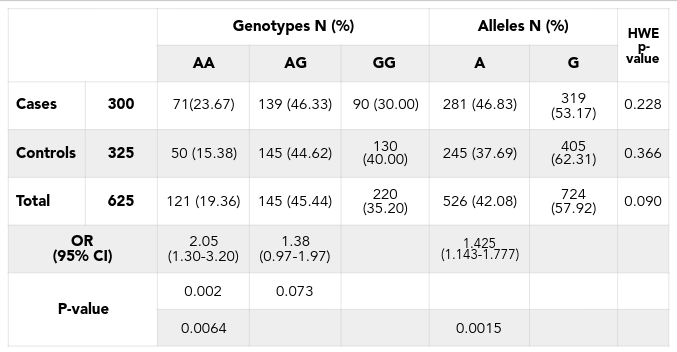
As a result of the good genotyping, the minor allele (allele A) of the SNP rs2046210 occupied 37.69% in control group and 62.31% in case group ( Table 1 ). Frequencies of three genotypes GG, AG and AA of this SNP in this population were 30.00%, 46.33% and 23.67% in case group and 40.00%, 44.62% and 15.38% in control group respectively ( Table 1 ). The Hardy-Weinberg equilibrium was calculated for total sample set and for each group of the set and the result showed that the frequencies of 3 genotypes are in equilibrium. Particularly, in total sample set, p-value is 0.09 (pTotal = 0.09), in control-group pControl = 0.336, and in case-group pCase = 0.228. This result indicated that the genotyping data is matched with the requirement for further association analysis.
Association analysis
The logistic regression analysis using STATA was applied for estimating the correlation between appearing of minor allele and the risk of getting the disease in the population. This statistic analysis showed that SNP rs2046210 is strongly associated with the disease, with very significant difference in allelic analysis [p = 0.0015, ORs (95% CI) = 1.43 (1.143 - 1.777)]. Genotypic analysis had also shown strong association between minor allele carriers and breast cancer with p = 0.0064. Homozygous genotypes of the minor allele (AA) have shown an increased risk of getting disease 2.05 folds compared to homozygous major allele (GG) with p = 0.002, ORs (95% CI) = 2.05[1.3 - 3.2] ( Table 1 ). Minor allele A is the risk allele, but in the heterozygous genotype, the person carried only 1 minor allele, seems not to be associated with breast cancer risk or weak association (OR [95% CI] = 1.38 [0.97 - 1.97], p = 0.073) ( Table 1 ). This indicated that the allele A has recessive effect in contribution to the breast cancer risk. The homozygous AA genotypes have shown strong effect (OR = 2.05) while the heterozygous AG genotypes have shown weaker effect (OR = 1.38).
The recessive effect of the allele A also is confirmed by the dominant analysis when all genotypes containing risk allele (AA and AG) is compared to the normal genotype (GG). Dominant analysis indicated that the risk allele carriers (women with the AG or AA genotypes), in general, increased 1.556 folds the risk of breast cancer (OR [95% CI] = 1.556 [1.116 - 2.168], p = 0.00892) ( Table 2 ).
However, the effect of both genotypes together was reduced with OR = 1.556 while the effect of AA genotype only has OR= 2.0556 in the additive ( Table 2 ).
Notably, recessive analysis does not show the complete recessive effect of the AG genotype. This recessive analysis demonstrated the weak effect of heterozygous AG genotypes to the risk of the diseases but has no effect on OR [95%CI] = 0.586 [0.392 - 0.877] ( Table 2 ).
Sample size and power estimation
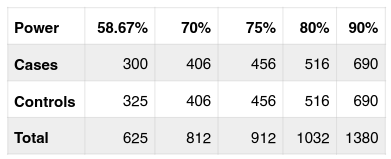
In genetic association study to detect the causal genes of complex human disease, the sample size with sufficient statistical power is critical to the success of the study. Based on the current sample size used in the study, strength of the study is calculated, and the result shows that with recent sample size (300 cases/ 325 controls) and the OR = 1.425 in the allelic analysis, the power of this study is 58.67%. It seems below the expectation. To get higher power up to 70% a sample size of 406 cases/406 controls should be investigated ( Table 3 ). With a bit bigger sample size, 456 cases/controls, the power of this study may go to 75%. And 80% or 90% power can be reached if the sample size is 516 cases/ controls or 690 cases/controls ( Table 3 ).
Discussion
From this study, the SNP rs2046210 has been confirmed as the risk allele A in Vietnamese population along with other populations such as Caucasian, Asian, European, Chinese and Japanese. Presence of allele A points to susceptibility to and increased risk of breast cancer, which is also consistent with those populations where the association is identified. The frequency of allele A in this study is 42.8% in the total sample set and 37.69% in control group, supported the data of previous studies in the Asian population, which was about 34.8 - 39.3% Chan et al., 2012Han et al., 2011. Specifically, in this study, 37.69% allele A in the control group was in the same range as the reported 34% in Caucasian descendants, 37.8% in Chinese (Hap Map-HCB) and 30.0% in Japanese (Hap Map - JTP) Mizoo et al., 2013. Distribution of this allele in Vietnamese which is the same as its distribution in other populations in Asia can be understood as they are in the same geography distribution and may share the same genetic distribution. Thus, the same genetic contribution to phenotype may also be the same. In fact, not only in Asian but this SNP was also found in European with nearly the same frequency (34%). However, in Africa, this ratio is relatively higher, around 69%, and this may lead to no association with breast cancer, while most of the previous studies in Chinese, Japanese, East-Asian, European, European-ancestry American, German women showed the association with the same frequency of risk allele Cai et al., 2011Stevens et al., 2011Yang et al., 2013.
The SNP rs2046210 was first identified as a risk variant for breast cancer among Chinese women by Zheng et al., 2009, after that several studies have replicated, and similar results were achieved. This polymorphism was significantly associated with the risk of breast cancer in different Asian populations, including Chinese with strong association [ORs (95% CI) = 1.35 (1.17 - 1.57), p = 4.1×10-5 for allelic analysis and [P for trend = 1.54×10−30] Lin et al., 2014. In Japanese women, the allele A has been strongly associated with breast cancer with [ORs (95% CI) = 1.37 (1.11 - 1.70), p = 0.05] Mizoo et al., 2013 or [ORs (95% CI) = 1.31 (1.13 - 1.52) and 1.37 (1.06 - 1.76) for the AG and AA genotypes, p for trend = 2.51×10−4] Cai et al., 2011. SNP rs2046210 had a population attributable risk of 18.9% plus an estimated 2.1% excess familial risk of breast cancer Zheng et al., 2009. As a result, the association between this SNP, in particular the allele A with breast cancer in Vietnamese in this study was also demonstrated with higher OR, 1.425 in this study comparing to 1.35 to 1.37 in Chinese and Japanese.
The power of this case/control study was estimated at 58.67%. It seems quite low comparing to the previous study in Chinese population Stevens et al., 2011. The power in Chinese study is around 77% with the sample size of 953 cases and 947 controls Stevens et al., 2011 while in this study, we only used 300 cases and 325 controls. With the power calculation, in case Vietnamese population, if the sample size is increased to about 500 cases/controls the strong association between SNP rs2046210 with the breast cancer will be confirmed with about 80%.
It is clear that with the sample size of 300 cases/325 controls, this association together with other studies on other populations, SNP rs2046210 is confirmed to be associated with breast cancer even with a bit low power. The association can be analysed in a bigger sample size to increase the power and increase the reliability. With this result and previous studies’ results, we believe that the allele A is the risk allele of breast cancer, not only for Vietnamese but also for other populations. Further functional studies are needed to confirm its function within the gene whether it alters the regulation of genes and leads to the breast cancer development. SNP rs2046210 is located 29 kb upstream of the first untranslated exon and 180 kb upstream of the transcription start site in the first exon of upstream of the estrogen receptor α (ESR1) gene on chromosome 6q25.1. This SNP is suspected to involve in regulating the expression of ESR1 gene suggesting its position within the binding site of a transcription factor Zheng et al., 2009. The allele A, a result of changing from allele G, may relate to some changes in the binding site of the transcription factor, thereby not allowing the transcription factor bind to the target DNA, then inhibit or induce the appropriate expression level of the gene ESR1. If that happens, an over-expression of ERα, which is the product of ESR1 gene might result and it relates to increase in the risk of breast cancer among people with A allele compared to people without A allele. As suggested by other studies, a higher level of ERα is related to the uncontrolled proliferation of breast cells Ellison-Zelski et al., 2009.
It is known that estrogen receptor positive (ER+ve) is one type of breast cancer in which the ESR 1 gene is expressed aberrantly high. This type of breast cancer can be considered as the most common one as it accounts for roughly 80% of human breast carcinomas Dunbier et al., 2011Russo and Russo, 2006. In this study, a population of breast cancer patients is selected as a representation for breast cancer disease in generally not separated in subtypes. Further analysis related to subtypes would be investigated to clarify this association. However, the association between SNP rs2046210 with breast cancer in this study indicated that the allele A of this SNP contributes to increase risk of the breast cancer development in Vietnamese in general. As it has been demonstrated to be associated with breast cancer in other populations Antoniou et al., 2011Long et al., 2010Zheng et al., 2009 or in a special subtype ER negative Yang et al., 2013 in the previous studies. Suggested that SNP rs2046210, particular allele A is a target marker for diagnosis breast cancer in the future.
Conclusion
In conclusion, with the selected sample set and the optimal HRM method, the SNP genotyping produced a highly reliable data. This case/control study confirmed that SNP rs2046210 is associated with the risk of breast cancer in Vietnamese population and allele A of this SNP contributed to increased risk of breast cancer susceptibility in a recessive effect.
Abbreviations
BC: Breast cancer; 95% CI: 95% confidence interval; Erα: estrogen receptor α; ESR1 gene: Estrogen receptor 1 gene; GWAS: Genome Wide Association Study; HRM: High Resolution Melting; HWE: Hardy-Weinberg equilibrium; NCBI : National Center for Biotechnology Information; OR: Odd ratio; PCR: Polymerase Chain Reaction; SNP: Single nucleotide polymorphism; WHO: World Health Organization
Author Contributions
NTTL contributed to study design, statistically analysed the data, and wrote the manuscript. BTN contributed to the study. NTNT, NDTG, PNH reviewed and edited the manuscript for intellectual content. Tran Van Thiep involved in samples collection. Nguyen Thi Hue oriented, gave important idea and revised the manuscript of this review. All authors gave final approval of the version to be published.
Ethical approval and informed consent
All patients gave informed consent and the studies were approved by the Local Ethics Committee of the Oncology Hospital of Ho Chi Minh City under the decision number 177/HĐĐĐ-CĐT, 18th November 2014.
References
-
A.C.
Antoniou,
C.
Kartsonaki,
O.M.
Sinilnikova,
P.
Soucy,
L.
McGuffog,
S.
Healey,
A.
Lee,
P.
Peterlongo,
S.
Manoukian,
B.
Peissel.
Common alleles at 6q25. 1 and 1p11. 2 are associated with breast cancer risk for BRCA1 and BRCA2 mutation carriers.. Human molecular genetics.
2011;
ddr226
.
-
Q.
Cai,
W.
Wen,
S.
Qu,
G.
Li,
K.M.
Egan,
K.
Chen,
S.L.
Deming,
H.
Shen,
C.-Y.
Shen,
M.D.
Gammon.
Replication and functional genomic analyses of the breast cancer susceptibility locus at 6q25. 1 generalize its importance in women of chinese, Japanese, and European ancestry. Cancer research.
2011;
71
:
1344-1355
.
-
M.
Chan,
S.
Ji,
C.
Liaw,
Y.
Yap,
H.
Law,
C.
Yoon,
C.
Wong,
W.
Yong,
N.
Wong,
R.
Ng.
Association of common genetic variants with breast cancer risk and clinicopathological characteristics in a Chinese population. Breast cancer research and treatment.
2012;
136
:
209-220
.
-
C.
DeSantis,
R.
Siegel,
A.
Jemal.
Breast cancer facts and figures 2013-2014. American Cancer Society.
2013;
:
1-38
.
-
A.K.
Dunbier,
H.
Anderson,
Z.
Ghazoui,
E.
Lopez-Knowles,
S.
Pancholi,
R.
Ribas,
S.
Drury,
K.
Sidhu,
A.
Leary,
L.-A.
Martin.
ESR1 is co-expressed with closely adjacent uncharacterised genes spanning a breast cancer susceptibility locus at 6q25. 1. PLoS Genet.
2011;
7
:
e1001382
.
-
S.J.
Dunstan,
N.T.
Hue,
B.
Han,
Z.
Li,
T.T.B.
Tram,
K.S.
Sim,
C.M.
Parry,
N.T.
Chinh,
H.
Vinh,
N.P.H.
Lan.
Variation at HLA-DRB1 is associated with resistance to enteric fever. Nature genetics.
2014;
46
:
1333-1336
.
-
D.F.
Easton,
K.A.
Pooley,
A.M.
Dunning,
P.D.
Pharoah,
D.
Thompson,
D.G.
Ballinger,
J.P.
Struewing,
J.
Morrison,
H.
Field,
R.
Luben.
Genome-wide association study identifies novel breast cancer susceptibility loci. Nature.
2007;
447
:
1087-1093
.
-
S.J.
Ellison-Zelski,
N.M.
Solodin,
E.T.
Alarid.
Repression of ESR1 through actions of estrogen receptor alpha and Sin3A at the proximal promoter. Molecular and cellular biology.
2009;
29
:
4949-4958
.
-
B.
Gold,
T.
Kirchhoff,
S.
Stefanov,
J.
Lautenberger,
A.
Viale,
J.
Garber,
E.
Friedman,
S.
Narod,
A.B.
Olshen,
P.
Gregersen.
Genome-wide association study provides evidence for a breast cancer risk locus at 6q22. 33.. Proceedings of the National Academy of Sciences.
2008;
105
:
4340-4345
.
-
W.
Han,
J.H.
Woo,
J.-H.
Yu,
M.-J.
Lee,
H.-G.
Moon,
D.
Kang,
D.-Y.
Noh.
Common genetic variants associated with breast cancer in Korean women and differential susceptibility according to intrinsic subtype. Cancer Epidemiology Biomarkers & Prevention.
2011;
20
:
793-798
.
-
N.T.
Hue,
N.D.H.
Chan,
P.T.
Phong,
N.T.T.
Linh,
N.D.
Giang.
Extraction of human genomic DNA from dried blood spots and hair roots. International Journal of.
2012;
Bioscience
:
Biochemistry and Bioinformatics 2, 21
.
-
D.J.
Hunter,
P.
Kraft,
K.B.
Jacobs,
D.G.
Cox,
M.
Yeager,
S.E.
Hankinson,
S.
Wacholder,
Z.
Wang,
R.
Welch,
A.
Hutchinson.
A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nature genetics.
2007;
39
:
870-874
.
-
Y.
Lin,
F.
Fu,
M.
Chen,
M.
Huang,
C.
Wang.
Associations of two common genetic variants with breast cancer risk in a Chinese population: a stratified interaction analysis. PloS one.
2014;
9
:
e115707
.
-
J.
Long,
Q.
Cai,
H.
Sung,
J.
Shi,
B.
Zhang,
J.-Y.
Choi,
W.
Wen,
R.J.
Delahanty,
W.
Lu,
Y.-T.
Gao.
Genome-wide association study in east Asians identifies novel susceptibility loci for breast cancer. PLoS Genet.
2012;
8
:
e1002532
.
-
J.
Long,
X.-O.
Shu,
Q.
Cai,
Y.-T.
Gao,
Y.
Zheng,
G.
Li,
C.
Li,
K.
Gu,
W.
Wen,
Y.-B.
Xiang.
Evaluation of breast cancer susceptibility loci in Chinese women. Cancer Epidemiology Biomarkers & Prevention.
2010;
19
:
2357-2365
.
-
T.
Mizoo,
N.
Taira,
K.
Nishiyama,
T.
Nogami,
T.
Iwamoto,
T.
Motoki,
T.
Shien,
J.
Matsuoka,
H.
Doihara,
S.
Ishihara.
Effects of lifestyle and single nucleotide polymorphisms on breast cancer risk: a case–control study in Japanese women. BMC cancer.
2013;
13
:
1
.
-
J.
Pei,
F.
Li,
B.
Wang.
Single nucleotide polymorphism 6q25. 1 rs2046210 and increased risk of breast cancer. Tumor Biology.
2013;
34
:
4073-4079
.
-
J.
Russo,
I.H.
Russo.
The role of estrogen in the initiation of breast cancer. The Journal of steroid biochemistry and molecular biology.
2006;
102
:
89-96
.
-
S.N.
Stacey,
A.
Manolescu,
P.
Sulem,
T.
Rafnar,
J.
Gudmundsson,
S.A.
Gudjonsson,
G.
Masson,
M.
Jakobsdottir,
S.
Thorlacius,
A.
Helgason.
Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor–positive breast cancer. Nature genetics.
2007;
39
:
865-869
.
-
K.N.
Stevens,
C.M.
Vachon,
A.M.
Lee,
S.
Slager,
T.
Lesnick,
C.
Olswold,
P.A.
Fasching,
P.
Miron,
D.
Eccles,
J.E.
Carpenter.
Common breast cancer susceptibility loci are associated with triple-negative breast cancer. Cancer research.
2011;
71
:
6240-6249
.
-
L.A.
Torre,
F.
Bray,
R.L.
Siegel,
J.
Ferlay,
J.
Lortet-Tieulent,
A.
Jemal.
Global cancer statistics, 2012. CA: a cancer journal for clinicians.
2015;
65
:
87-108
.
-
WHO.
Early detection of cancer.. In Cancer (World Health Organization).
2016
.
-
C.T.
Wittwer,
G.H.
Reed,
C.N.
Gundry,
J.G.
Vandersteen,
R.J.
Pryor.
High-resolution genotyping by amplicon melting analysis using LCGreen. Clinical chemistry.
2003;
49
:
853-860
.
-
Z.
Yang,
J.
Shen,
Z.
Cao,
B.
Wang.
Association between a novel polymorphism (rs2046210) of the 6q25 1 locus and breast cancer risk. Breast cancer research and treatment.
2013;
139
:
267-275
.
-
W.
Zheng,
J.
Long,
Y.-T.
Gao,
C.
Li,
Y.
Zheng,
Y.-B.
Xiang,
W.
Wen,
S.
Levy,
S.L.
Deming,
J.L.
Haines.
Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25. 1. Nature genetics.
2009;
41
:
324-328
.
-
X.
Zhou,
Y.
Gu,
D.-n.
Wang,
S.
Ni,
J.
Yan.
Eight functional polymorphisms in the estrogen receptor 1 gene and endometrial cancer risk: a meta-analysis. PloS one.
2013;
8
:
e60851
.
Comments
Downloads
Article Details
Volume & Issue : Vol 3 No 11 (2016)
Page No.: 973-984
Published on: 2016-11-18
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 4869 times
- Download PDF downloaded - 1320 times
- View Article downloaded - 41 times
 Biomedpress
Biomedpress