Abstract
Introduction: With 170 million chronic hepatitis C virus (HCV) cases worldwide, HCV is considered a major life-threatening pathogen. HCV is a crucial causative of liver cirrhosis and hepatocellular carcinoma. Tumor necrosis factor-alpha (TNF-alpha) is thought to be a mediator in the development of viral hepatitis. Because HCV is epidemic in Egypt, this study aimed to characterize the distribution of TNF-alpha gene promoter polymorphisms and their relation to TNF-alpha expression in HCV patients.
Methods: Four promoter polymorphisms; −1031T/C, −863C/A, −857C/T, and −308G/A, were studied by restriction fragment length polymorphism in a population of Egyptian HCV patients.
Results: Compared to healthy subjects, none of these polymorphisms were associated with HCV infection. The wild-type −1031T, −863A, −857C, and −308G alleles were highly prevalent in the studied population. Sequencing the promoter region spanning the four studied polymorphisms in some subjects did not reveal any difference in the nucleotide variance pattern, compared with the TNF-alpha reference sequence. Relative TNF-alpha mRNA expressions in HCV patients and healthy subjects were statistically indifferent.
Conclusion: Since previous studies confirmed an increase in TNF-alpha level in case of viral infections, this study focuses on mechanisms of post-transcriptional and posttranslational modifications of TNF-alpha gene in HCV patients, which decode the genetic factors linked to HCV infection and severity.
Introduction
The Hepatitis C virus (HCV) is the major cause of liver cirrhosis and hepatocellular carcinoma (HCC), which is the most common causative of liver transplantation in several countries 1. Globally, about 170 million people are chronically infected with HCV2. Chronic liver inflammation develops by the aid of various mediators, which may act as cofactors in carcinogenesis 3. Among these mediators, the tumor necrosis factor-α (TNF-α) is a critical pathogenic mediator in the development of viral hepatitis. TNF-α is required for the propagation of liver cells in case of liver damage. It also mediates hepatotoxicity 4.
Various types of cells produce TNF-α. To become biologically active, it binds to two target cell receptors: TNFR1 and TNFR2 5. TNF-α-encoding gene is located on chromosome 6 in the class III region of the major histocompatibility complex. Its promoter region contains several polymorphisms, the most common of which are −1031T/C, −863C/A, −857C/A, −376G/A, −308G/A, and −238G/A. Some of these genetic polymorphisms (1031T/C, −376G/A, −308G/A, and −238G/A) were found to affect TNF-α expression levels 6.
TNF-α ‒308G/A polymorphism was linked to the pathogenesis and advancement of chronic hepatitis C (CHC) 7,8. Dogra et al. 9 reported a significant association between TNF-α ‒308GG genotype and HCV infection in patients when compared with healthy individuals. Dai et al. 10 reported that detecting of TNF-α ‒308A allele before administering combination therapy might be useful for predicting the treatment response, especially in difficult-to-treat HCV patients.
TNF-α was reported as a carcinogenesis cofactor 3 with a potential role in cancer pathogenesis 11, and its level elevated in HCC patients 12. According to Yang et al. 11 meta-analysis, TNF-α ‒308G/A polymorphism was assumed to confer a higher risk of HCC, especially in the Asian population. Xiao et al. 13 meta-analysis revealed that increased HBV-HCC risk was associated with TNF-α ‒308AA and TNF-α ‒238AA, and with TNF-α ‒863CA genotypes. In some Chinese populations, TNF-α ‒308G/A polymorphism was associated with HCC susceptibility 14.
In Egypt, HCV is an epidemic with a prevalence of 14.7% 15. Approximately 75-85% of HCV-infected persons will progress to chronic infection and risk developing HCC 1. This study aims to evaluate the distribution of TNF-α gene polymorphisms in HCV-infected patients in the Egyptian population and explore whether any of these polymorphisms are linked to HCV-infection and TNF-α expression.
Subjects - methods
Subjects
Blood samples were collected from 100 subjects (50 HCV RNA-positive patients and 50 healthy subjects). The study was ethically authorized by our institute’s board (No. 5/2/2/1 and 5/2/2/8), and all subjects gave their written consent before samples were collected. All HCV-positive patients were Hepatitis B virus (HBV)-negative and rheumatoid factor-negative, and the healthy subjects were negative for both of HCV, HBV, and rheumatoid factors.
Genotyping
Genomic DNA was extracted from blood samples by Gene JETTM Genomic DNA Purification Kit (#K0722, Thermo Scientific, Waltham MA, USA), according to the manufacturer’s instructions. All extracted DNA samples were genotyped by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) to detect ‒1031T/C (rs1799964), ‒863C/A (rs1800630), ‒857C/T (rs1799724), and ‒308 G/A (rs1800629) polymorphisms in the promoter of TNF-α gene according to Skoog et al. 16. Genotyping of the ‒1031T/C polymorphism was performed by amplifying a DNA fragment with the forward primer 5′-TATGTGATGGACTCACCAGGT-3′ and the reverse primer 5′-CCTCTACATGGCCCTGTCTT-3′. The amplified fragment was cut with the restriction enzyme BbsI (#ER1011, Thermo Scientific, Waltham MA, USA). Genotyping for the ‒863C/A and ‒857C/T polymorphisms were performed by amplifying fragments with the common forward primer 5′-GGCTCTGAGGAATGGGTTAC-3′ and the reverse primer 5′-CTACATGGCCCTGTCTTCGTTACG-3′ for ‒863C/A, and the reverse primer 5′-CCTCTACATGGCCCTGTCTAC-3′ for ‒857C/T. The two types of amplified fragments were digested with the restriction enzyme TaiI (#ER1141, Thermo Scientific, Waltham MA, USA). Genotyping of the ‒308G/A polymorphism was performed by amplifying a DNA fragment with the forward primer 5′-GAGGCAATAGGTTTTGAGGGCCAT-3′ and the reverse primer 5′-GGGACACACAAGCATCAAG-3′. The amplified fragment was digested with the restriction enzyme NcoI (#ER0571, Thermo Scientific, Waltham MA, USA).
Each PCR was a 25 µL reaction mixture, containing 25 ng of gDNA, ten pmoles of each primer, 12.5 µL of Hot Start PCR master mix (#K1051, Thermo Scientific, Waltham MA, USA). PCRs were conducted using a thermocycler (Multigene, Labnet, Edison NJ, USA) under the following conditions: 95°C × 10 min, (95°C × 30 sec, 60°C × 30 sec, 72°C × 30 sec) × 35 cycles, and 72°C × 5 min.
Digestion reactions were done in 30 µL reaction mixtures according to the enzyme manufacturer’s instructions at the recommended temperature for 16 hrs. Digested products were run side by side with equivalent amounts of undigested PCR products on 5% agarose gels for 1 hr in TBE buffer, except for ‒1031T/C polymorphism where the products were run on 3% gels. Gels were stained with ethidium eromide and documented in the Photo Doc-IT Imaging System (UVP, Upland CA, USA).
Quantitation of TNF-α mRNA expression
Total RNA was isolated from all samples by Gene JET™ RNA extraction kit (#K0731, Thermo Scientific, Waltham MA, USA) according to the manufacturer’s instructions. cDNA was produced from pure RNA preparations by means of RevertAid™ First Strand cDNA Synthesis Kit (#K1631, Thermo Scientific, Waltham MA, USA), according to the recommended instructions. cDNA preparations were used immediately or stored at ‒20°C when necessary.
Quantitative Real Time PCR for TNF-α was performed by Maxima SYBR Green PCR Master Mix (#K0251, Thermo Scientific, Waltham MA, USA) in 20 µL reaction mixtures, following the recommended instructions. TNF-α expression was quantitated using the forward primer 5′-CTTCTCCTTCCTGATCGTGG-3′ and the reverse primer 5′-CCCTGGGGAACTCTTCCCTCT-3′ 17. β-actin was quantitated as a housekeeping gene, using the forward primer 5′-CCTTCTACAAATGAGCTGCGT-3′ and the reverse primer 5′-CCTGGATAGCAACGTACATG-3′. Reactions were cycled in CFX96 Real Time PCR detection system (Bio-Rad, CA, USA) as follows: 95°C × 10 min, (95°C × 15 sec, 60°C × 20 sec, 72°C × 30 sec) × 40 cycles. Data were analyzed by the ΔCt method.
DNA sequencing
DNA isolated from 10 subjects was amplified by PCR as mentioned above using the forward primer 5′-TATGTGATGGACTCACCAGGT-3′ and the reverse primer 5′-ATCTGGAGGAAGCGGTAGTG-3′. PCR products were purified by The MEGAquick-spin Plus Total Fragment DNA Purification Kit (#MQP17289, Intron, Seol, South Korea) according to the manufacturer’s instructions and sequenced by the standard Sanger method at Macrogen sequencing facility (Macrogen, Seol, South Korea). Chromatograms were checked, and the sequences were aligned to the TNF-α gene reference sequence (GenBank: NG_007462).
Statistical analysis
Results were statistically evaluated using the SPSS package version 22 (IBM Inc., New York, USA). TNF-α expression was statistically assessed by Student's t-test, and P value ⩽0.05 was considered statistically significant. Genotype and allele distributions were tested by the Chi-squared test. The distribution of TNF-α promoter genotypes in each group was checked for deviations from Hardy–Weinberg equilibrium using a web-based tool 18.
Results
Genotype frequencies
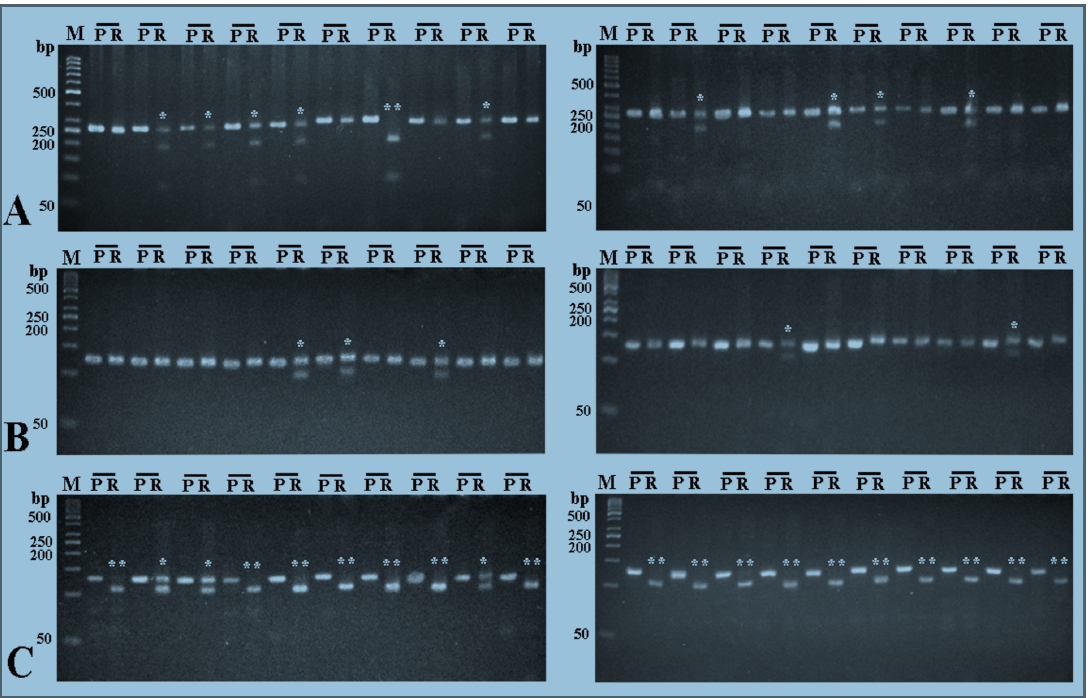
To investigate whether there is an association between TNF-α gene polymorphisms and the risk of infection with HCV, DNA was extracted from the blood of 50 HCV RNA positive-patients and 50 healthy subjects, and TNF-α gene promoter was investigated by RFLP technique for the presence of 4 different polymorphisms; ‒1031T/C, ‒863C/A, ‒857C/T, and ‒308G/A.
For ‒1031T/C polymorphism, the homozygous wild TT genotype was detected in 50% of the HCV patients and 60% of the healthy subjects, while the heterozygous TC genotype was detected in 44% of HCV patients and 34% of healthy subjects. The homozygous CC genotype was detected in 6% of HCV patients and, also, 60% of healthy subjects (Figure 1A&B).
For ‒863C/A polymorphism, the homozygous wild CC genotype was detected in 68% of HCV patients and 66% of the healthy subjects, while the heterozygous genotype was detected in 32% of HCV patients and 34% of the healthy subjects. The homozygous AA genotype was not detected in either HCV patients or the healthy subjects (Figure 1C&D).
With respect to ‒857C/T polymorphism, the homozygous wild CC genotype was detected in 74% of HCV patients and 76% of the healthy subjects, while the heterozygous genotype was detected in 24% of both HCV patients and the healthy subjects. The homozygous TT genotype was detected in 2% of HCV patients and was not detected in any of the healthy subjects (Figure 1E&F).
Regarding ‒308G/A polymorphism, the homozygous wild GG genotype was detected in 88% of HCV patients and 84% of the healthy subjects, while the heterozygous GA genotype was detected in 12% of HCV patients and 16% of the healthy subjects. The homozygous AA genotype was not detected in both the HCV patients and healthy subjects (Figure 1G&H).
There were no significant differences in the observed frequency of any genotype between HCV patients and the healthy subjects (Table 1 ). The distributions of all genotypes among these two groups were both in Hardy–Weinberg equilibrium (Table 2).
| Polymorphism | Genotype | HCV patients | Healthy subjects | χ 2 | p-value | ||
| N=50 | % | N=50 | % | ||||
| TT | 25 | 50 | 30 | 60 | 1.01 | 0.42 | |
| ‒1031 T/C | TC | 22 | 44 | 17 | 34 | 1.05 | 0.41 |
| CC | 3 | 6 | 3 | 6 | 0.00 | 1.00 | |
| CC | 34 | 68 | 33 | 66 | 0.45 | 1.00 | |
| ‒863 C/A | CA | 16 | 32 | 17 | 34 | 0.45 | 1.00 |
| AA | 0 | 0 | 0 | 0 | - | - | |
| CC | 37 | 74 | 38 | 76 | 0.05 | 1.00 | |
| ‒857 C/T | CT | 12 | 24 | 12 | 24 | 0.00 | 1.00 |
| TT | 1 | 2 | 0 | 0 | 1.01 | 1.00 | |
| GG | 44 | 88 | 42 | 84 | 0.33 | 0.77 | |
| ‒308 G/A | GA | 6 | 12 | 8 | 16 | 0.33 | 0.77 |
| AA | 0 | 0 | 0 | 0 | - | - | |
| Locus | Genotype | HCV patients | Healthy subjects | ||
| Frequency | HWE χ 2 | Frequency | HWE χ 2 | ||
| ‒1031 | TT, TC, CC | 25, 22, 3 | 0.42 | 3, 17, 30 | 0.08 |
| ‒863 | CC, CA, AA | 34, 16, 0 | 1.81 | 0, 17, 33 | 2.10 |
| ‒857 | CC, CT, TT | 37, 12, 1 | 0.00 | 38, 12, 0 | 0.93 |
| ‒308 | GG, GA, AA | 44, 6, 0 | 0.20 | 42, 8, 0 | 0.38 |
Frequencies of allele carriage
Rates of allele carriage in HCV patients or healthy subjects were calculated (Table 2). The ‒1031T allele was carried by 94% of either HCV patients or healthy subjects, and the ‒1031C allele was carried by 50% of HCV patients and 40% of healthy subjects, respectively. The ‒863C allele was carried by 100% of either HCV patients or healthy subjects, and the ‒863A allele was carried by 32% of HCV patients and 34% of healthy subjects. The ‒857C allele was carried by 98% of HCV patients and 100% of the healthy subjects; the ‒857T allele was carried by 26% of HCV patients and 24% of the healthy subjects. The ‒308G allele was carried by 100% of both HCV patients and the healthy subjects, and the ‒308A allele was carried by 12% of HCV patients and 16% of the healthy subjects. The differences in these allele frequencies were statistically insignificant (Table 3).
| Allele | HCV patients | Healthy subjects | χ 2 | p-value | ||
| N=50 | % | N=50 | % | |||
| ‒1031 T | 47 | 94 | 47 | 94 | 0.00 | 1.00 |
| ‒1031 C | 25 | 50 | 20 | 40 | 0.72 | 0.50 |
| ‒863 C | 50 | 100 | 50 | 100 | 0.00 | 1.00 |
| ‒863 A | 16 | 32 | 17 | 34 | 0.03 | 1.00 |
| ‒857 C | 49 | 98 | 50 | 100 | 0.02 | 1.00 |
| ‒857 T | 13 | 26 | 12 | 24 | 0.04 | 1.00 |
| ‒308 G | 50 | 100 | 50 | 100 | 0.00 | 1.00 |
| ‒308 A | 6 | 12 | 8 | 16 | 0.30 | 0.78 |
Frequencies of allele carriage in the studied Egyptian population (HCV patients plus healthy subjects) were summarized in Table 4. The wild-type ‒1031T, ‒863C, ‒857C, and ‒308G alleles were highly distributed (p=0.00) in the Egyptian population than the non-wild alleles.
| Allele | Population (N=100) % | χ 2 | p-value |
| ‒1031 T | 94 | 26.47 | 0.00 |
| ‒1031 C | 45 | ||
| ‒863 C | 100 | 50.00 | 0.00 |
| ‒863 A | 33 | ||
| ‒857 C | 99 | 64.00 | 0.00 |
| ‒857 T | 25 | ||
| ‒308 G | 100 | 90.73 | 0.00 |
| ‒308 A | 14 |
Partial sequencing of TNF-α gene promoter
To confirm the PCR-RFLP genotyping results and compare the sequence of the TNF-α gene promoter in Egyptians with the reference TNF-α gene sequence (GenBank: NG_007462), the promoter region (‒1063 to ‒230), spanning the four studied polymorphisms were sequenced. Sequence results confirmed the polymorphic patterns drawn from the PCR-RFLP results. Additionally, the nucleotide variance in this promoter region of 10 studied subjects (5 healthy subjects and 5 HCV patients) was identical to the reference sequence, except for the known polymorphisms −1031T/C, −863C/A, −857C/A, −376G/A, −308G/A, and −238G/A (Table 5).
| Reference sequence | Healthy #1 | Healthy #2 | Healthy #3 | Healthy #4 | Healthy #5 | HCV #1 | HCV #2 | HCV #3 | HCV #4 | HCV #5 | Total | |
| -1,031 | T | C | C | C | C | C | C | 6 | ||||
| -863 | C | M | A | A | 3 | |||||||
| -857 | C | T | 1 | |||||||||
| -376 | G | A | A | R | 3 | |||||||
| -308 | G | R | R | A | A | 4 | ||||||
| -238 | G | R | A | A | R | A | R | A | 7 | |||
| Total differences | 2 | 1 | 3 | 4 | 4 | 2 | 4 | 2 | 2 | 24 |
Quantitation of TNF-α mRNA expression
Expression of TNF-α mRNA in HCV patients and the healthy subjects was evaluated by real-time PCR. The relative TNF-α mRNA expression in HCV patients and the healthy subjects was statistically indifferent (p=0.21) (Figure 2).
Discussion
TNF-α has been considered the fundamental pathogenic mediator in various liver conditions, including hepatic fibrogenesis and fibrosis progression in chronic liver disease 19. It has been shown that TNF-α aids in activating hepatic stellate cells and transforming them into an activated myofibroblast-like phenotype, which leads to accumulation of extracellular matrix and fibrosis development 20. Besides, TNF-α is thought to prompt the production of other fibrogenic factors, such as tumor growth factor-β, IL-1, and IL-6 21.
TNF-α levels increase in patients with HBV infection 22, and TNF-α production has been linked to the polymorphisms in its promoter region 23. Some polymorphisms in TNF-α promoter were correlated with the severity of liver disease in HBV patients and thought to influence the susceptibility to chronicity, which outcomes as cirrhosis, or HCC 24.
This study investigated the polymorphisms in 4 consecutive loci in the TNF-α promoter; ‒1031T/C, ‒863C/A, ‒857C/T, and ‒308G/A, in a population of HCV infected Egyptian patients. Compared to the healthy subjects, the observed frequency of all genotypes and alleles in HCV patients and the healthy subjects were statistically indifferent. The distributions of all genotypes among HCV patients and the healthy subjects were both in the Hardy–Weinberg equilibrium. None of the four studied polymorphisms were associated with HCV infection. Additionally, the wild-type ‒1031T, ‒863A, ‒857C, and ‒308G alleles were highly common among the studied Egyptian population. The sequence of the promoter region, spanning the four studied polymorphisms did not reveal any nucleotide difference from the GenBank TNF-α reference sequence, except for the known polymorphisms.
Studies correlating susceptibility to HCV infection with certain TNF-α promoter polymorphisms are limited and conflicting. A significant association between TNF-α ‒308GG genotype and HCV infections in Indian patients compared with healthy individuals was reported 9. Talaat et al. 25 reported a significantly higher TNF-α ‒308G allele frequency in HCV-infected Egyptian patients, compared with healthy controls, and a higher frequency of TNF-α ‒308A allele in healthy control individuals. In contrast to this, TNF-α ‒308A allele was significantly associated with HCV infection among Egyptian patients 26. Except for ‒308G/A, no other TNF-α polymorphisms were investigated in Egyptian HCV patients.
Several studies correlated TNF-α polymorphisms with the chronicity of HCV infection, the susceptibility to HCC development and response to therapy. TNF-α ‒308G/A polymorphism was involved in the pathogenesis and advancement of CHC in Caucasian patients 7,8. Both TNF-α ‒308G/A and ‒857C/T polymorphisms were significantly associated with acute viral hepatitis in Indians 27. These results are inconsistent with several studies which did not notice any association between TNF-α ‒308G/A polymorphisms and severity of fibrosis in CHC Caucasian Spanish patients 28, Tunisian patients 29, and Indian patients 9. The severity of CHC infection was not associated with TNF-α ‒308G/A polymorphisms in Caucasian patients and Irish patients 30,31. TNF-α ‒308A allele was associated with increased susceptibility to severe HCV recurrence after transplant 7 and with a 3.2-fold increased risk of cirrhosis for Caucasian patients, having chronic HCV infection 8. However, no association was recorded between TNF-α ‒308G/A polymorphisms and susceptibility to chronic HCV infection, or viral persistence in Taiwanese patients 32. Similarly, meta-analysis studies 33,34 stated the absence of such an association. Besides, no association between ‒308G/A polymorphisms and liver cirrhosis risk was noticed in both Caucasian and Asian patients 35. In HCV-infected Pakistani patients, sustained virological response was not affected by TNF-α ‒308G/A polymorphism 36.
Generally, data of the studies on TNF-α genetic polymorphisms have been reported to vary from one study to another, and such variation was related to the differences in the ethnic origins or the number of the individuals involved in the study6. Additionally, the difference in applied techniques, as well as the data interpretation, could contribute to the aforementioned discrepancy.
A lack of association between the four studied polymorphisms and HCV infection in the current study has prompted determining TNF-α expression in HCV patients. Results did not show any significant difference in the relative TNF-α mRNA expression between HCV patients and the healthy subjects, thus supporting the lack of association between the studied genotypes/alleles and HCV infection.
It is documented that genetic polymorphisms, certain promoter sequences, and allelic variants are associated with low or high cytokine production in vitro and in vivo 23,37. Polymorphisms at the positions ‒1031, ‒863, ‒857, ‒308, and -238 of TNF-α gene promoter were differentially associated with TNF-α production in different populations 38.
Individuals with the genotypes ‒308AA or AG were reported to have higher levels of serum TNF-α and, in contrast to this, individuals with the genotypes ‒863AA or CA were noted to have lower TNF-α level 39. Although the allele ‒308A is commonly associated with increased production of TNF-α 23,40, some other studies 41,42 demonstrated that ‒308G/A polymorphism does not have a significant effect on TNF expression. In Chinese populations, the genotypes ‒308GA and AA were correlated with decreased serum TNF-α level 43.
TNF-α ‒863C/A polymorphism was reported to impact binding of nuclear proteins to the promoter region of the TNF-α gene, causing variation in the serum TNF-α level. Specifically, the ‒863A allele had significantly less serum TNF-α quantity 16. In the current study, the genotypes ‒308AA or AG were less frequent in both HCV patients and healthy subjects; however, the ‒863A allele showed a 100% occurrence frequency in both groups.
Previous studies indicated that both of the ‒857T and ‒857C alleles caused high levels of transcriptional activity of TNF-α mRNA and protein with a stronger effect for ‒857T allele 44. The ‒857T allele caused a 1.7-fold higher level of transcription activity, compared to the ‒857C allele 45. In the current study, the allele ‒857T was highly frequent in both HCV patients and healthy subjects.
Studies on the effect of ‒1031T/C polymorphism on TNF-α expression are very limited. In Chinese populations, the genotype ‒1031TC and CC was correlated with decreased serum TNF-α level. Chinese ‒308G/A, ‒857C/T, and ‒1031T/C combination carriers had lower serum TNF-α level than the carriers of the corresponding wild type 43.
TNF-α promoter polymorphisms do affect TNF-α expression level, but the activation and abundance of other molecules that directly or indirectly interact with certain promoter sequences must affect the expression of TNF-α 6.
Identification of the genetic factors associated with HCV susceptibility and severity is fundamental for understanding the mechanism, provoking the development of chronic hepatitis into cirrhosis and HCC. Such Identification also facilitates implementing suitable treatment strategies. Due to the absence of absolute quantitation of TNF-α level in this study, it is uncertain whether TNF-α level was high or low in both HCV patients and the healthy subjects of the studied Egyptian population. Although, most of the previous studies that discussed the role of TNF-α support the notion of an increase in TNF-α level in case of viral infections. The similarity in TNF-α mRNA expression in both HCV patients and the healthy subjects in the Egyptian population rises several essential questions. Firstly, whether the mechanisms of the post-transcriptional and posttranslational modifications of the TNF-α gene in HCV patients, compared to the healthy subjects are the same? Secondly, whether the ethnic nature predisposes the Egyptian population to some TNF-α-related diseases like viral infections, autoimmune diseases, and cancers if this cytokine is left uncontrolled? Additional studies on TNF-α expression in a large population of Egyptians may provide certain answers.
Conclusion
This study investigated the distribution of 4 TNF-α gene promoter polymorphisms (‒1031T/C, ‒863C/A, ‒857C/T, and ‒308G/A) and their relation to TNF-α expression in HCV-infected Egyptian patients. None of these polymorphisms were associated with HCV infection. The promoter region spanning these polymorphisms in some subjects was similar to the TNF-α GenBank reference sequence. No significant difference was detected in the relative TNF-α mRNA expression between HCV patients and healthy subjects. Further studies are needed to determine whether the post-transcriptional and posttranslational modifications of TNF-α in HCV patients and healthy subjects are different.
Abbreviations
CHC: chronic hepatitis C
HBV: hepatitis B virus
HCC: hepatocellular carcinoma
HCV: hepatitis C virus
PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism
TNF-α: Tumor necrosis factor-α
Competing Interests
The author declares that there is no conflict of interest in this study.
References
-
Chen
S.L.,
Morgan
T.R.,
The natural history of hepatitis C virus (HCV) infection. Int J Med Sci.
2006;
3
(2)
:
47-52
.
View Article PubMed Google Scholar -
Organization
World Health,
Hepatitis C, fact sheet N 164. 2017
.
-
Coussens
L.M.,
Werb
Z.,
Inflammation and cancer. Nature.
2002;
420
(6917)
:
860-7
.
View Article PubMed Google Scholar -
Bradham
C.A.,
Plümpe
J.,
Manns
M.P.,
Brenner
D.A.,
Trautwein
C.,
Mechanisms of hepatic toxicity. I. TNF-induced liver injury. Am J Physiol.
1998;
275
(3)
:
387-92
.
PubMed Google Scholar -
Locksley
R.M.,
Killeen
N.,
Lenardo
M.J.,
The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell.
2001;
104
(4)
:
487-501
.
View Article PubMed Google Scholar -
El-Tahan
R.R.,
Ghoneim
A.M.,
El-Mashad
N.,
TNF-α gene polymorphisms and expression. Springerplus.
2016;
5
(1)
:
1508
.
View Article PubMed Google Scholar -
Rosen
H.R.,
Lentz
J.J.,
Rose
S.L.,
Rabkin
J.,
Corless
C.L.,
Taylor
K.,
Donor polymorphism of tumor necrosis factor gene: relationship with variable severity of hepatitis C recurrence after liver transplantation. Transplantation.
1999;
68
(12)
:
1898-902
.
View Article PubMed Google Scholar -
Yee
L.J.,
Tang
J.,
Herrera
J.,
Kaslow
R.A.,
van Leeuwen
D.J.,
Tumor necrosis factor gene polymorphisms in patients with cirrhosis from chronic hepatitis C virus infection. Genes Immun.
2000;
1
(6)
:
386-90
.
View Article PubMed Google Scholar -
Dogra
G.,
Chakravarti
A.,
Kar
P.,
Chawla
Y.K.,
Polymorphism of tumor necrosis factor-α and interleukin-10 gene promoter region in chronic hepatitis C virus patients and their effect on pegylated interferon-α therapy response. Hum Immunol.
2011;
72
(10)
:
935-9
.
View Article PubMed Google Scholar -
Dai
C.Y.,
Chuang
W.L.,
Chang
W.Y.,
Chen
S.C.,
Lee
L.P.,
Hsieh
M.Y.,
Tumor necrosis factor alpha promoter polymorphism at position -308 predicts hepatitis C virus response to combination therapy. J Infect Dis.
2006;
193
(1)
:
98-101
.
View Article PubMed Google Scholar -
Yang
Y.,
Luo
C.,
Feng
R.,
Bi
S.,
The TNF-α, IL-1B and IL-10 polymorphisms and risk for hepatocellular carcinoma: a meta-analysis. J Cancer Res Clin Oncol.
2011;
137
(6)
:
947-52
.
View Article PubMed Google Scholar -
Morsi
M.I.,
Hussein
A.E.,
Mostafa
M.,
El-Abd
E.,
El-Moneim
N.A.,
Evaluation of tumour necrosis factor-alpha, soluble P-selectin, gamma-glutamyl transferase, glutathione S-transferase-pi and alpha-fetoprotein in patients with hepatocellular carcinoma before and during chemotherapy. Br J Biomed Sci.
2006;
63
(2)
:
74-8
.
View Article PubMed Google Scholar -
Xiao
Q.,
Fu
B.,
Chen
P.,
Liu
Z.Z.,
Wang
W.,
Ye
Q.,
Three polymorphisms of tumor necrosis factor-alpha and hepatitis B virus related hepatocellular carcinoma: A meta-analysis. Medicine (Baltimore).
2016;
95
(50)
:
e5609
.
View Article PubMed Google Scholar -
Li
Y.,
Ou
C.,
Shu
H.,
Zhao
H.,
Zhu
B.,
The ERCC1-4533/8092, TNF-α 238/308 polymorphisms and the risk of hepatocellular carcinoma in Guangxi Zhuang populations of China: case-control study. Medicine (Baltimore).
2016;
95
(44)
:
e5217
.
View Article PubMed Google Scholar -
Mohamoud
Y.A.,
Mumtaz
G.R.,
Riome
S.,
Miller
D.,
Abu-Raddad
L.J.,
The epidemiology of hepatitis C virus in Egypt: a systematic review and data synthesis. BMC Infect Dis.
2013;
13
(1)
:
288
.
View Article PubMed Google Scholar -
Skoog
T.,
van't Hooft
F.M.,
Kallin
B.,
Jovinge
S.,
Boquist
S.,
Nilsson
J.,
A common functional polymorphism (C\textemdash>A substitution at position -863) in the promoter region of the tumour necrosis factor-alpha (TNF-alpha) gene associated with reduced circulating levels of TNF-alpha. Hum Mol Genet.
1999;
8
(8)
:
1443-9
.
View Article PubMed Google Scholar -
Mousa
A.K.,
Ghoneim
A.M.,
El-Mashad
N.B.,
El-Ghobashy
A.,
TNF% UNKNOWN UNICODE CHARACTER 02011 (NON-BREAKING HYPHEN) α genetic polymorphisms and its expression in Egyptian rheumatoid arthritis patients. Am J Life Sci..
2014;
2
(4)
:
234-40
.
View Article Google Scholar -
Rodriguez
S.,
Gaunt
T.R.,
Day
I.N.,
Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol.
2009;
169
(4)
:
505-14
.
View Article PubMed Google Scholar -
Richardson
M.M.,
Powell
E.E.,
Barrie
H.D.,
Clouston
A.D.,
Purdie
D.M.,
Jonsson
J.R.,
A combination of genetic polymorphisms increases the risk of progressive disease in chronic hepatitis C. J Med Genet.
2005;
42
(7)
:
e45-45
.
View Article PubMed Google Scholar -
Özdemir
B.H.,
Bilezikçi
B.,
Haberal
M.,
Hepatic stellate cells in hepatitis C patients: relationship with the development of interstitial fibrosis in renal allografts. Transplant Proc.
2009;
41
(7)
:
2838-40
.
View Article PubMed Google Scholar -
Balkwill
F.,
Cancer and the chemokine network. Nat Rev Cancer.
2004;
4
(7)
:
540-50
.
View Article PubMed Google Scholar -
Zhang
G.,
Li
Z.,
Han
Q.,
Li
N.,
Zhu
Q.,
Li
F.,
Altered TNF-α and IFN-γ levels associated with PD1 but not TNFA polymorphisms in patients with chronic HBV infection. Infect Genet Evol.
2011;
11
(7)
:
1624-30
.
View Article PubMed Google Scholar -
Wilson
A.G.,
Symons
J.A.,
McDowell
T.L.,
McDevitt
H.O.,
Duff
G.W.,
Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci USA.
1997;
94
(7)
:
3195-9
.
View Article PubMed Google Scholar -
Du
T.,
Guo
X.H.,
Zhu
X.L.,
Li
J.H.,
Lu
L.P.,
Gao
J.R.,
Association of TNF-alpha promoter polymorphisms with the outcomes of hepatitis B virus infection in Chinese Han population. J Viral Hepat.
2006;
13
(9)
:
618-24
.
View Article PubMed Google Scholar -
Talaat
R.M.,
Esmail
A.A.,
Elwakil
R.,
Gurgis
A.A.,
Nasr
M.I.,
Tumor necrosis factor-alpha -308G/A polymorphism and risk of hepatocellular carcinoma in hepatitis C virus-infected patients. Chin J Cancer.
2012;
31
(1)
:
29-35
.
PubMed Google Scholar -
Pasha
H.F.,
Radwan
M.I.,
Hagrass
H.A.,
Tantawy
E.A.,
Emara
M.H.,
Cytokines genes polymorphisms in chronic hepatitis C: impact on susceptibility to infection and response to therapy. Cytokine.
2013;
61
(2)
:
478-84
.
View Article PubMed Google Scholar -
Singhal
S.,
Kohaar
I.,
Bharadwaj
M.,
Shukla
D.K.,
Das
B.C.,
Kar
P.,
Association of tumor necrosis factor-alpha gene promoter polymorphisms with acute viral hepatitis in the Indian population. Dig Dis Sci.
2010;
55
(4)
:
1106-12
.
View Article PubMed Google Scholar -
Romero-Gómez
M.,
Montes-Cano
M.A.,
Otero-Fernández
M.A.,
Torres
B.,
Sánchez-Muñoz
D.,
Aguilar
F.,
SLC11A1 promoter gene polymorphisms and fibrosis progression in chronic hepatitis C. Gut.
2004;
53
(3)
:
446-50
.
View Article PubMed Google Scholar -
Bouzgarrou
N.,
Hassen
E.,
Gabbouj
S.,
Schvoerer
E.,
Ben Mami
N.,
Triki
H.,
Lack of effect of tumor necrosis factor-alpha -308 G/A polymorphism on severity of liver fibrosis in Tunisian hepatitis C virus (HCV)-infected patients. Gastroenterol Clin Biol.
2010;
34
(4-5)
:
297-304
.
View Article PubMed Google Scholar -
Höhler
T.,
Kruger
A.,
Gerken
G.,
Schneider
P.M.,
Meyer zum Büschenfelde
K.H.,
Rittner
C.,
Tumor necrosis factor alpha promoter polymorphism at position -238 is associated with chronic active hepatitis C infection. J Med Virol.
1998;
54
(3)
:
173-7
.
View Article PubMed Google Scholar -
Barrett
S.,
Collins
M.,
Kenny
C.,
Ryan
E.,
Keane
C.O.,
Crowe
J.,
Polymorphisms in tumour necrosis factor-alpha, transforming growth factor-beta, interleukin-10, interleukin-6, interferon-gamma, and outcome of hepatitis C virus infection. J Med Virol.
2003;
71
(2)
:
212-8
.
View Article PubMed Google Scholar -
Dai
C.Y.,
Chuang
W.L.,
Lee
L.P.,
Chen
S.C.,
Hou
N.J.,
Lin
Z.Y.,
Associations of tumour necrosis factor alpha promoter polymorphisms at position -308 and -238 with clinical characteristics of chronic hepatitis C. J Viral Hepat.
2006;
13
(11)
:
770-4
.
View Article PubMed Google Scholar -
Chen
Y.,
Pei
J.,
An assessment of a TNF polymorphic marker for the risk of HCV infection: meta-analysis and a new clinical study design. Infect Genet Evol.
2009;
9
(6)
:
1356-63
.
View Article PubMed Google Scholar -
He
J.,
Pei
X.,
Xu
W.,
Wang
C.,
Zhang
X.,
Wu
J.,
The relationship between tumor necrosis factor-α polymorphisms and hepatitis C virus infection: a systematic review and meta-analysis. Ren Fail.
2011;
33
(9)
:
915-22
.
View Article PubMed Google Scholar -
Chen
D.,
Liu
J.L.,
Liu
Y.,
Zhu
J.,
Wang
S.W.,
Lack of an association between -308G>A polymorphism of the TNF-α gene and liver cirrhosis risk based on a meta-analysis. Genet Mol Res.
2011;
10
(4)
:
2765-74
.
View Article PubMed Google Scholar -
Abbas
Z.,
Moatter
T.,
Hussainy
A.,
Jafri
W.,
Effect of cytokine gene polymorphism on histological activity index, viral load and response to treatment in patients with chronic hepatitis C genotype 3. World J Gastroenterol.
2005;
11
(42)
:
6656-61
.
View Article PubMed Google Scholar -
Gibson
A.W.,
Edberg
J.C.,
Wu
J.,
Westendorp
R.G.,
Huizinga
T.W.,
Kimberly
R.P.,
Novel single nucleotide polymorphisms in the distal IL-10 promoter affect IL-10 production and enhance the risk of systemic lupus erythematosus. J Immunol.
2001;
166
(6)
:
3915-22
.
View Article PubMed Google Scholar -
Hananantachai
H.,
Patarapotikul
J.,
Ohashi
J.,
Naka
I.,
Krudsood
S.,
Looareesuwan
S.,
Significant association between TNF-alpha (TNF) promoter allele (-1031C, -863C, and -857C) and cerebral malaria in Thailand. Tissue Antigens.
2007;
69
(3)
:
277-80
.
View Article PubMed Google Scholar -
Huang
Y.C.,
Chang
W.C.,
Shan
Y.H.,
Lin
C.Y.,
Wang
C.L.,
Dai
C.Y.,
Toxic Metals Increase Serum Tumor Necrosis Factor-α Levels, Modified by Essential Elements and Different Types of Tumor Necrosis Factor-α Promoter Single-nucleotide Polymorphisms. Epidemiology.
2017;
28
:
113-20
.
View Article PubMed Google Scholar -
Kroeger
K.M.,
Carville
K.S.,
Abraham
L.J.,
The -308 tumor necrosis factor-α promoter polymorphism effects transcription. Mol Immunol.
1997;
34
(5)
:
391-9
.
View Article PubMed Google Scholar -
Brinkman
B.M.,
Zuijdeest
D.,
Kaijzel
E.L.,
Breedveld
F.C.,
Verweij
C.L.,
Relevance of the tumor necrosis factor alpha (TNF alpha) -308 promoter polymorphism in TNF alpha gene regulation. J Inflamm.
1995-1996;
46
(1)
:
32-41
.
PubMed Google Scholar -
Uglialoro
A.M.,
Turbay
D.,
Pesavento
P.A.,
Delgado
J.C.,
McKenzie
F.E.,
Gribben
J.G.,
Identification of three new single nucleotide polymorphisms in the human tumor necrosis factor-alpha gene promoter. Tissue Antigens.
1998;
52
(4)
:
359-67
.
View Article PubMed Google Scholar -
Cui
G.,
Wang
H.,
Li
R.,
Zhang
L.,
Li
Z.,
Wang
Y.,
Polymorphism of tumor necrosis factor alpha (TNF-alpha) gene promoter, circulating TNF-alpha level, and cardiovascular risk factor for ischemic stroke. J Neuroinflammation.
2012;
9
(1)
:
235
.
View Article PubMed Google Scholar -
Kimura
K.,
Takayanagi
R.,
Yokoyama
H.,
Yamada
Y.,
Effects of tumor necrosis factor α-857C/T polymorphism on the expression of tumor necrosis factor α. APMIS.
2016;
124
(8)
:
669-74
.
View Article PubMed Google Scholar -
Higuchi
T.,
Seki
N.,
Kamizono
S.,
Yamada
A.,
Kimura
A.,
Kato
H.,
Polymorphism of the 5'-flanking region of the human tumor necrosis factor (TNF)-α gene in Japanese. Tissue Antigens.
1998;
51
(6)
:
605-12
.
View Article PubMed Google Scholar
Comments
Downloads
Article Details
Volume & Issue : Vol 6 No 5 (2019)
Page No.: 3156-3165
Published on: 2019-05-30
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
- HTML viewed - 4290 times
- Download PDF downloaded - 1240 times
- View Article downloaded - 0 times
 Biomedpress
Biomedpress




