Abstract
Obesity and overweight have become a global problem that development of various coronary artery diseases (CAD), such as myocardial infarction, atherosclerosis and congestive heart failure. Effective diagnosis is needed for effective treatment and prevention, particularly in healthy overweight subject. Platelets is an important component of hemostatic balance, that maintains the coagulation physiology. Platelets are involved in different pathological events such as thrombosis and CAD in various inflammatory conditions. There are evidences that highlight an important role of miRNA in regulation of gene expression profiling in platelets. Current hypothesis has shown the likelihood of using miRNAs as diagnostic markers in the event of CAD. This review article describes the association between overweight/obesity and platelets activation in elucidating the gene expression profiling in platelet miRNAs in CAD patient. The application of platelet miRNAs as predictive markers in overweight/obese individuals may become a marginal milestone in the history of this diet-related disorder treatment.
Introduction
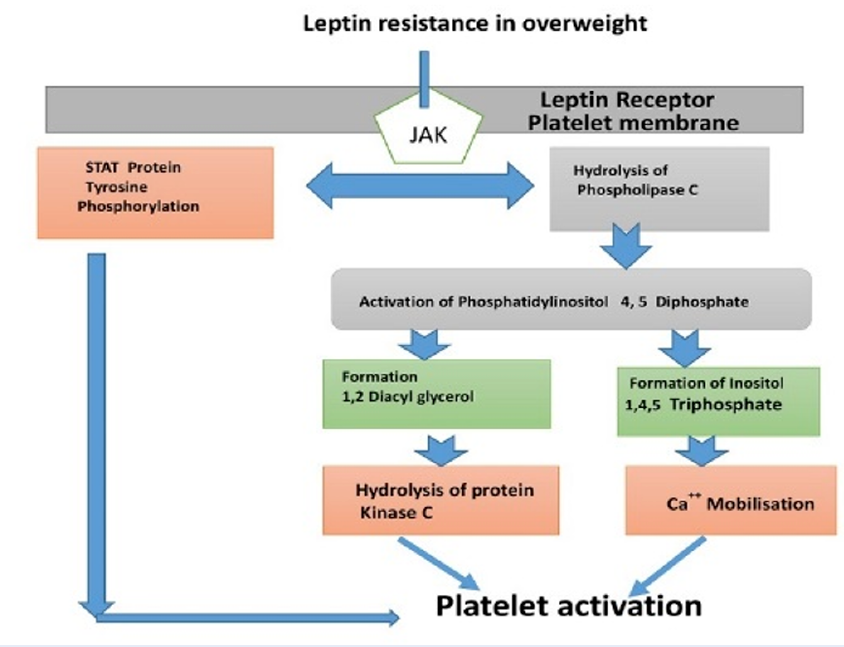
Overweight is a major public health issue, which affects conspicuous portion of the world population. It constitutes a risk factor for metabolic events that predispose an individual to diabetes, hypertensions and atherosclerosis. Overweight has been recently linked to low grade chronic inflammatory diseases, like type 2 diabetes, cardiovascular disease and cancer. However, acute inflammatory responses are important for defensive and homeostatic mechanisms of healing, repair and tissue regeneration1. Chronic inflammatory conditions include hypertension, diabetes, cardiovascular disorders (CVDs) and cancer2. A report by World Health Organization (WHO) in 2016 showed that over 1.9 billion adults above 18 were overweight worldwide, and 650 million adults were obese. These figures show, that 39 % of adults ≥ 18 years of age (representing 40 % women and 39 % men) were overweight, while 13 % of world adult population (representing 15 % women and 11 % men) were obese. Thus, from 1975 to 20163, the global prevalence of overweight and obesity cases has nearly tripled. Epidemiological evidence and clinical studies have clearly demonstrated, that overweight predisposed an individual to an increased incidence of thrombotic occlusion, or atherosclerosis that correlated with epicardial fat thickness4 and visceral obesity5. Endothelial dysfunction, monocytes recruitment, inflammation and platelet activation are associated with dyslipidemia. Studies showed that leptin resistance could be a possible candidate that links obesity with cardiovascular diseases6,7,8. Meta-analysis data by Coronary Heart Prevention of West Scotland Studies suggested, that leptin could induce CHD independently9. Although, leptin receptor (ObRb) on platelet membrane can signal angiogenesis, regulate bone formation, accelerate vascular endothelial injuries and further enhance platelet aggregation via platelet leptin receptor10,11 (Figure 1).
Activated platelet releases cytokines, chemokines, hemostatic factor, adhesion protein, antigen receptor, mitogens and platelet miRNA12. Platelet miRNA gene expression pattern has been demonstrated to be more accurate and precise, compared to mRNA expression pattern in differentiation and characterizing various diseases (such as atherosclerosis, myocardial infarction and cancers 13). Thus, this review article describes the relationship between overweight, leptin resistance, platelet activation and platelets miRNAs expression profiling, as well as to advocates the usefulness of miRNAs as predictive biomarkers of CAD in healthy overweight subjects.
The association between platelet activation and atherosclerosis in overweight
Overweight can be referred as a leptin resistance condition. Leptin is a product of adiponectin from ob gene14, that signals the energy stores level via receptor in central nervous system. Leptin regulatesactivity of the hypothalamic nuclei, responsible for appetite and energy homeostasis. This hormone is anti-overweight, with its level decreasing during fasting and increasing after overfeeding in order to maintain energy status. Thus, leptin resistance in overweight subjects may signify the relationship between leptin concentration and overweight15. Multiple evidences have demonstrated that this hormone could engage in many pathophysiological pathways that could result in leptin resistance in different cells, arterial thrombosis formation16, platelet activation and aggregation17, arterial hypertension18 and vascular response to inflammation19,20. Other studies have shown that leptin is a prognosticator of atherosclerosis, myocardial infarction, stroke and coronary artery episode which is independent of body fat21.
The overall ability of this hormone to increase human platelet activation has been suggested in OB/OB mice and healthy control subjects11,21. However, Ozata et al. did not observe leptin on epinephrine, collagen and ADP platelet activation in both obese and overweight healthy subjects, and in leptin-deficient individuals22. Therefore, the notion of presence of leptin receptor on platelet membrane, signaling the prothrombotic episode of leptin resistance is still controversial. However, the association between platelet activation in overweight/obesity with dyslipidemia and endothelial dysfunction, triggers human platelet activation and aggregation, thus further enhancing the risk of atherosclerosis events23. Furthermore, studies have shown that obesity triggers platelet activation via release of an inflammatory mediator that activates multiple internal signaling networks, including platelet miRNA24. The role of miRNA (miR) on leptin resistance has been demonstrated in several gene expression profiles. MiRNA plays a modulation role in different metabolites. Thus, the exact effect of platelet miRNA on leptin in hypothalamic center remains elusive. The up-regulation of miR-200b and miR-200a were observed in hypothalamus center of obese-deficient ob/ob mice, and down-regulation of miR-200b and miR-200a were reported after leptin treatment in the hypothalamus25. Reduced level of miR-200b and miR-200a in the hypothalamic center induced the expression of both insulin and leptin receptors that signal the reduction of fat deposit and further restored insulin function in the liver 26. These miRNAs were present in activated platelets, suggesting that up regulation of miR-200a and miR-200b from activated platelets in overweight/obesity was altered (Table 1). Such alteration influenced leptin and insulin receptor signaling pathways in the hypothalamic nuclei25 which could serve as a target for therapeutic intervention in obesity/overweight individuals.
Platelet activation and protein synthesis
Platelets are made up of three secretory granules: the α-granules, lysosomes and dense granules27. The content of these granules is released into plasma after platelet activation, which may enhance and promote elevation levels of pro-atherogenic proteins. This includes growth factors (TGF-β, PDGF, bFGF and EGF), adhesion proteins (fibronectin, fibrinogen, P-selectin, thrombospondin, vWF, vitronectin, receptor complex glycoproteinIba-V-IX (GPIba-V-IX,), collagen receptor GP VI and GPIIb-IIIa), epithelial neutrophil activating protein 78 (ENA-78), chemokines CXC chemokine ligand 4 (CXCL4), RANTES (CCL5), platelet factor 4 (PF4), coagulation factors (factor V, XI, XIII), α2-antiplasmin, PAI-1 (plasminogen activator inhibitor), TFPI (tissue factor pathway inhibitor), protein S, antithrombin, plasminogen, cytokine-like factors (CD40L, β-thromboglobulin and IL-1β) and miRNA28 (Figure 2).
Regulatory and b iogenesis of platelet miRNA
MiRNAs are a short class of non-coding endogenous RNAs, that post-regulate gene expression transcriptionally via either translational mRNA degradation, or repression. MiRNAs play crucial roles in controlling biochemical functions and mechanisms of different types of cells, including cell differentiation, developmental timing, tumorigenesis, proliferation, apoptosis as well as thrombosis and platelet functions. Similarly, miRNA itself as a regulatory element, is coordinatively modulated by multifarious effectors when carrying out basic functions, such as miRNA editing, single-nucleotide polymorphism, circadian clock and methylation. Platelet miRNAs act together in order to fine-tune and regulate a wide variety of this molecules in form of mRNA in different cellular functions. Those include aggregation, cell adhesion, proliferation, activation, chemotaxis, coagulation, proteolysis and cell survival. During these events, the mRNA is synthesized from DNA transcription and further translated into protein. Certain specific regions of these mRNA molecules are not normally translated into proteins. These regions include the 5′ untranslated region (5’ UTR), 5′ cap, poly-A tail and 3′ untranslated region (3' UTR). 3′ UTR often contains regulatory site that influences and regulates the post-transcriptional gene expression profiling29. Britton and Davidson, (1969)30 postulated that "activator" mRNA transcript may work to turn on and off genes as predicted by Watson-Crick base pairing to region, located within genes31. Three major forms of sRNAs regulation in plants and animals were identified, which include small interfering RNAs (siRNA), miRNAs (miRNA) and piwi-interacting RNAs (piRNA). Landry et al., 200932 found, that the platelet miRNA was not usually translated into protein, but it regulated mRNA by annealing to the recognition site on 3’ UTR of mRNA, coding gene sequence of 2-8 nucleotide, complementary to miRNA seed region33. This pairing was based on Watson-Crick nucleotide base pairing, such as miRNA-mRNA. The hybridization of these complexes does not require full complementary homology34 and complete homolog usually leads to degradation of a target mRNA, while partial pairing homology is a translational repression. Therefore, miRNA influences degradation to regulate mRNA by switching the functional gene transcript on and off, while translational repression of miRNA regulates the function of the protein expression34,35. This functional role of miRNA presumes a wide range of miRNA-mRNA interactions, following either convergent (many miRNA-Single targets), or divergent (single miRNA-many target mRNA) interaction. This results in robust pathways that regulate the synthesis of protein in a cell35.
Currently, the nature and extent of the involvement of platelet miRNAs in a non-(protein)-coding ribonucleic acids (RNAs) in physiology and pathological processes (in both plant and animals) has become clearer35. More than 280 miRNAs have been recognized and characterized from human nucleated platelets31,35. The miRNAs biosynthesis in biological systems is strictly regulated.
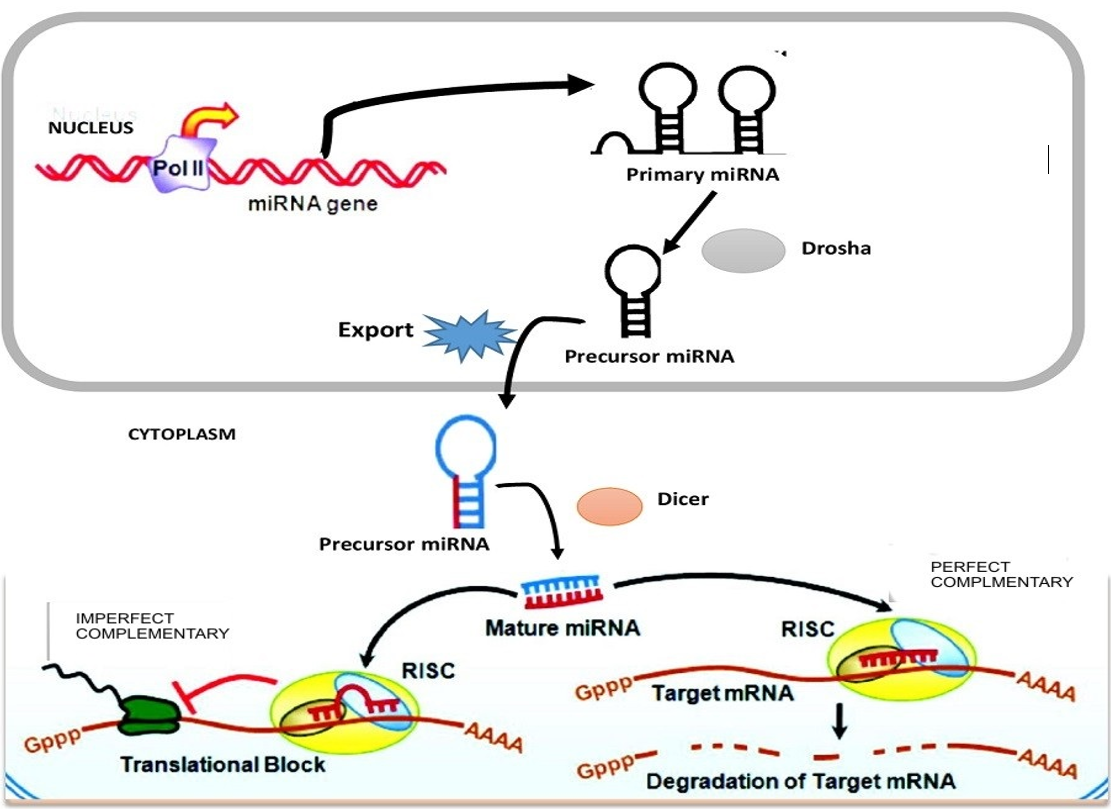
The biogenesis of platelet miRNAs began with transcription of the gene in non-coding region of miRNA using RNAs poly-III from the genome of megakaryocytes, harboring miRNA gene in either intergenic or intronic at promoter site. The first primary miRNA as a non-coding transcript is known as pri-miRNA, which acts upon by RNA poly III (called “Drosha”), that bounds to the microprocessor subunits DGCR-8 (di-george syndrome critical region-8) to produce a small length hairpin called pre-miRNA stemloop with 80-110 nucleotides base in length. Exporting-5 assists the nuclear export of pre-miRNA into the cytoplasm36. The presence of pre-miRNAs in the cellular cytoplasm is identified by ribonuclease (RNase) or Dicer enzymes which is a part of transactivation response binding protein (TRBP) complex. The complex Dicer/TRBP cleaves the pre-miRNAs loop to release a short length of double stranded miRNA, such as miRNA/miRNA, * or duplex37. The duplex (such as miRNA/miRNA) are transported into the RNA Induced Silencing Complex (RISC)38,39 and Helicase enzymes are used to unwind the double stranded miRNA into a single 22 nucleotides base, which is released as matured miRNA. The 2nd strand miRNA, known as passenger strand is thought to be digested by the RISC complex substrate. Argonauts (Ago) protein family will assemble in RISC and provide stability and protection to the mature miRNA strand against RNase enzymes activities and will further guide the matured miRNA to its target 3′-untranslated region (UTR) of mRNA transcripts40 (Figure 3 ). However, the inverse modulation of platelet miRNA profiling during adipose tissue development in overweight and obesity is important for understanding miRNA dysregulation in adipose tissue of both obese humans and mice. Such modulation enhances chronic inflammation observed in obesity subject with insulin resistance41. Thus, a single nucleotide base change of mature and pre-miRNA may drive the emerging of new miRNA by influencing the biological function of these cells40.
Pathological role of platelet miRNA in coronary artery disease
The mutations or single-nucleotide polymorphism (SNP) that occur in the biogenesis of both mRNA and miRNA can be classified as follows:
(1) Mutations, that affect biogenesis enzymes of miRNA, or the promoter region of mRNA.
(2) Mutations, that affect the 3’ UTR region of the mRNA.
(3) Mutations that affect the miRNA seed region.42
Mutation or SNP that affects miRNA biogenesis enzymes
SNP in the processing regions can be categorized as pri-miRNA, pre-miRNA, mature miRNA sequences and mRNA biogenesis machinery (promoter region). In fact, gene variations may influence either miRNA hairpin and biogenesis proteins, or enzymes in the processing accuracy43.
Mutation in the miRNA seed region
SNP in the seed region of miRNA may reduce the binding strength between miRNA and mRNA target sites, which will influence hundreds of gene expressions44.
Mutation in the mRNA 3’ UTR-region
Approximately 180,000 mutations or SNP found within the 3' UTR region mRNA were demonstrated with its corresponding mature miRNA 2,600 sequences. These were found in database sequence45. Mutations in the seed region of miRNA and 3’ UTR region of mRNA are almost identical in terms of variations and regulation of miRNA-mRNA* complex, affecting functions and disease vulnerability42. However, further molecular mechanism underlying disease-associated with 3’ UTR SNPs in mRNA still require further investigation46. For example, SNP meta-analysis of 8,120 patients and 8,364 controls identified four different SNPs in the following miRNAs (miRNA-149, miRNA-196a2, miRNA-499 and miRNA-146a), which increased patients CAD vulnerability47. Other articles have demonstrated that miR-146a was upregulated in CAD 47,48 due to the change of G to C in pre-miRNA. That affected the expression of mature miRNA-146a in CAD patients49. That finding was confirmed by Wang et al. and showed, that C allele was greater than G allele in gene that predisposed individuals to CAD 50,51. However, in type 2 diabetes patients with ischemic stroke, the miR-146a was downregulated (Table 1) 52. Other abundant miRNAs found in platelets miRNA were miR-223 and miR-126. In addition, miR-223, miR-107, miR- 495, miR-96 and miR-200b were actively involved in various platelet functions, such as platelet release, adhesion and aggregation,53 as summarized in Table 2.
miRNA as biomarkers
The fact of discovery of miRNA as a stable molecule in plasma and serum is surprising despite the level of RNase enzymes activities 54. The expression of miRNA in tissue, or organ-specific, and its release into the plasma in response to tissue injury is considered to be a potential biomarker for different diseases55. The hypothesis that miRNA in blood cells could be used as diagnostic parameter in different diseases was first postulated by Mitchell et al.. He showed, that miRNA remained highly stable in both plasma and serum even after prolonged storage at room temperature, or repeated cycles of freezing and thawing. For example, tumor-derived miRNA could easily be identified in plasma sample from cancer patient with standard laboratory procedure56,57,54. The diagnostic role of circulating miRNA has been evaluated not only in cancer, but also in other clinical disorders, such as hepatic disease58, heart failure59 and diabetes60. Few studies have claimed platelets in obesity and overweight healthy individuals as the culprit factor, responsible for coronary artery disease61. The application of specific platelet miRNAs as markers for platelet activation will be a marginal milestone in the history of this diet-related disorder.
Platelet- miRNA in endothelial and vascular smooth muscle (VSCM) cells in coronary artery disease (CAD)
Large scale studies, presented by Nagalla et al. and Landry et al. found, that thirty different platelet miRNAs modulate platelet and endothelial angiogenesis (angio-miRs)32,35. These miRNAs, (miR-21, miR-200, miR-210 and miR-126), are well known to play critical roles in the vessel and capillary formation. The regulation of angiogenesis by miRNA in the endothelial cell was confirmed, using Dicer-knockdown mice experiment, demonstrating the essential role of Dicer in miRNA biogenesis62,63. Furthermore, Dicer gene deletion was shown to cause early death of mice during the embryonic stage, due to impaired angiogenesis64. In addition, Dicer knockdown of vascular smooth muscle cells VSMC specific caused the late embryonic death due to internal bleeding65, suggesting that platelet miRNA was an integral part of vascular development. Therefore, clusters of miR-221 and miR-222 are the most abundant and well distributed miRNAs across these three cells(platelet, endothelial and VSMC)66,67. Both miR-221 and miR-222 function as pro-inflammatory inhibitor of angiotensin II and reversed leucocytes adhesion (in vivo and in vitro)68, suggesting a possible role of these miRNA clusters in CAD dysregulation. The endothelial-enriched miR-92a has been proposed as a possible therapeutic target after treatment with anti-miR-92a oligonucleotide.Such treatment improved the formation of blood vessel and recovery of cardiovascular disorder in acute myocardial infarction in mice67. MiR-143/145 is the most abundant miRNA found in VSCM and is well characterized as part of the same bi-cistronic cluster. These miRNAs target multiple mRNAs to influence VSCM differentiation and simultaneously reduce proliferation68. The delivery of miR-145 by lentiviral in endothelial and VSCM inhibits monocyte/macrophages recruitment and infiltration, thus reducing inflammation and limiting plaque formation. These results suggest a new therapeutic target in order to decrease atherosclerotic progression and increase plaque atrial stability69.
Role of platelet miRNA as a marker in coronary artery disease (CAD)
Previous reports showed that more than 80 different diseases were associated with dysregulation, or mutation in miRNAs70. The mechanisms of platelet activation in various thrombotic diseases were well established71. The accurate etiology of miRNA in activated platelet in overweight and obesity is not clear, but obesity is always a risk factor for atherothrombotic episodes71. Landry et al. 2009 demonstrated that the platelets and megakaryocyte miRNAs had 219 different types of miRNA in platelet expression patterns and profiles72. The authors observed three most abundant miRNAs in human platelets; miR-19a, let-7c, and miR-223. Binding of miRNA-223 to 3' UTR region of P2Y12 mRNA receptor in HEK293 cell line repressed P2Y12 gene and decreased the activities of both platelet and megakaryocyte. However, dysregulation of miRNA-233 was observed in hyperreactive platelet, leading to overexpression of miRNA-22373. Thus, miR-223, miR-197 and miR-126 were involved in platelet hyperreactive and endovascular inflammation, stretching the application of these miRNAs as predictive biomarkers for diagnosis of CAD (Table 1)73.
| Platelet miRNA | Disease | Group | Methods | Reference |
| ↓mir-223 | Diabetes mellitus type 2 patients without ischemic stroke | Human/7 diabetes, 8 control | qRT-PCR | 52 |
| ↓miR942, ↓20a ↑miR146a-5p* | Acute coronary syndrome (NSTEMI) | Human/13 CAD, 13 non- | qRT- PCR | 74 |
| ↑ miR127-3p ↓miR185-5p* | Acute coronary syndrome (STEMI) | Human/44 CAD, 22 non-CAD | qRT-PCR | 75 |
| ↑miR454* ↑miR545:9.1↓ miR-12801* | Premature coronary artery disease | Human/12 premature CAD, 12 controls | Microarray confirmed by qRT-PCR | 53 |
| ↑miR-144, ↓miR-146a* | Diabetes mellitus type 2 patients with ischemic stroke | HUMAN/6 ischemic stroke ,8 controls | Microarray qRT-PCR | 52 |
| ↑miR340* ↑miR624* | Mature coronary artery disease | Human/40 CAD, 40 controls | Microarray Confirmed by qRT-PCR | 53 |
CAD, Coronary Artery Disease; qRT-PCR, Real-Time Quantitative Reverse Transcription PCR
In 2013, Osman and Fälker discovered 281 transcripts, of which 228 were mature miRNAs and 53 were pre-miRNAs. Six miRNAs: miR-339-3 p, miR-15 a, miR-365, miR-495, miR-361-3 p and miR-98, were upregulated, or downregulated in hyperreactive platelets28. The level of expression pattern, or characteristic of miRNAs in platelets were associated with a procoagulant (such as thrombin stimulation). Nagalla et al., 2011 demonstrated that there were 284 miRNA transcripts in human platelets, by which 74 were differentially expressed, based on the platelet reactivity35. Seven miRNA expression profiles (miR-320b, miR-190, miR-320d, miR-19b, miR-34b, miR-320c and miR-320a) were shown to have a strong relationship with the degree of platelet response to adrenaline35. In fact, the most abundant expressed platelets miRNA is miR-223, followed by miR-126 57. Others were identified as miR-200b, miR-107, miR-96 and miR- 49553. All of these miRNAs were involved in platelet hyperreactivity (such as platelet activation, adhesion and aggregation, as summarized in Table 2).
| Platelet miRNA | Target protein(mRNA) | Function | Pathophysiology | Implication | Ref | |
|---|---|---|---|---|---|---|
| 1. | miRNA-223 | P2Y12 receptorfor ADP | Regulate platelet Hyperreactive | miRNA-233: Dysregulation of miRNA-233 will results in hyperreactive platelet, leading to upregulation miRNA-223↑ | miR-223 and miR-197 are platelet activation miRNA involved in vascular inflammation and have been shown as markers in the diagnosis of CAD. | 73 , 76 |
| 2. | miRNA-200b | cAMP-dependent PKA | Keep platelet in hyporeactive state | miRNA-200b: Dysregulation in obesity/overweight subjects lead to release of plate αlpha- and dense granules that harbor inflammatory molecules in platelet. miRNA-200b inhibits endothelial angiogenesis upregulates↑ | Overexpression of miR-200b in platelet activation may be used as a diagnostic maker of CAD | 77 , 78 |
| 3 | miR-107 | CLOCK/Bmal1 | vWF gene is regulated by CLOCK/Bmal1 complex in normal physiological state | miRNA-107: Dysregulationincreased the plasma level of vWF↑ | Association with a prothrombotic state that promote CAD | 79 |
| 4. | Member of miR-17-92 cluster (miR-17) | Fibronectin | Fibronectin aids in the formation of stable arterial thrombi at the site of endothelialinjury | Inhibition of fibronectin by miR-17 leads to impeded platelet coagulation and wound healing and is downregulated ↓ | miR-17-92 and its family were dysregulated in CAD | 80 , 81 |
| 5 | miR29a*/miR-409-3p* and fibrinogen | FGA, FGB,and FGG) | Fibrinogen functions as hemostatic plug formation, | miR-29 reduce the level of mRNA genes such as FGG, FGB and FGA. Dysregulation of these miRNAs has been linked with cardiac fibrosis ↓ | miR-29 family are grouped with MI-regulated member | 82 , 83 |
| 6. | miR-96 and miR-15 | VAMP8 | miR-96 and miR-15 regulate VAMP8.VAMP8 aids in platelet activation, secretion of α-granules and platelet function. | SNPs in the 3’ untranslated region ( UTR) of VAMP8 mRNA resulted in VMP8 mRNA dysregulation and is upregulated ↑ | The consequencerisk of myocardial infarction | 84 |
| 7. | miR-495 | Kelch protein | Platelet activation uses Kelch protein for actin and filament organization during platelet function | KLHL5 mRNA is required for platelet activation and miR-495 regulates the Kelch-like protein at 3’ UTR region. Repression of this protein in megakroyocytes leads to platelet hyporeaction | ? | 85 |
| miR-126 present in both platelet and endothelial cell | VCAM-1 | The target of miRNA-126 is VCAM mRNA, and the suppression of VCAM leads to decreasing infiltration of leucocytes into the vascular endothelial cell | Mutation of miR-126 resulted in the expression of TNF-α that stimulate VCAM-1 expression and increase leukocyte adherence to endothelial ↓ | Promotes CAD | 86 , 87 | |
| 9 | miR-210. | ephrina3 receptor (efna3) and Tyrosine protein and Phosphatase(ptp1b) | The repression of the two molecules (ptp1b and efna3) by miRNA-210 enhances angiogenic property and endothelial angiogenesis | Mutation lead to downregulation of miR-210↓ | Angiogenesis after MI | 88 |
| miR-21 | Rho kinase, SPROYTY2, RhoB,BMPRII, SOD2 and PTEN | All possess anti-angiogenic functions.. | Increased expression of miR-21 inhibits angiogenesis of endothelial cells by downregulating several pro-angiogenicmiR-21↑ | Atherosclerosis, CAD, apoptosis and neoangiogenes | 89 , 90 , 91 |
Plé and colleagues in 2012 detected more than 492 different mature miRNA transcripts in active platelets74,75,92. A total of 15 novel miRNAs were identified from human platelet: miR-103, miR-140, miR-24, miR-185, miR-223, miR-23, miR-320, miR-25, miR-21, miR-26, miR-191, miR-423, miR-101, miR-199 and let-786. This finding suggested a possible relationship between the platelet activation and the miRNA modification, which may an induce agonist-specific platelet function.
Jeanine et al., 2014 also reported the expression profile of platelet miRNA in patients with acute coronary syndrome after a series of diagnostic investigations. In blood sample of STEMI patients, miR185-5p and miR186-5p were downregulated, while miR221-3p and miR127-3p were upregulated in platelets. On the other hand, in blood samples of NSTEMI patients, miR942 and miR20a-5p were downregulated, whereas miR146a-5p and miR483-5p were upregulated in platelets (Table 2)74. Thus, in body circulation, the pathophysiology of platelet miRNA can exhibit clear characteristic differences of expression profiling in various platelet disorders. This condition however is dangerous if urgent measure is not taken, since the risk of heart failure (HF) in such case is eminent. Patients with systolic heart failure were found to have different platelet miRNA expression profiles, as compared to control subjects56. Platelets miRNA-150 expression level decreased more than three-fold in blood platelets of patients with heart failure, secondary to atrial fibrillation (Table 2) 75.
In addition, the study, conducted by Duan et al., 2014 showed, that the expression of platelet miRNA-223 and miRNA-146a in patients with ischemic stroke and diabetes mellitus was significantly higher, compared to healthy donors. The expression level of these two platelet miRNAs was suggested to correlate with platelet activation (Table 2)52.
Conclusion
The gene expression and bioinformatic analysis procedures have become the state-of-the-art methods for diagnosing a number of human diseases. Several studies have reported different miRNA expression profiles in platelets. The application of these miRNAs for diagnostic purpose in overweight healthy subjects will be a milestone in the history of treatment of this diet-related disorder. Six notable candidates of platelet miRNAs have been discovered and proposed for diagnostic accuracy in CAD upregulation: ↑ miR483-5p* ↑ miR146a-5p* ↑miR340* ↑miR624*, ↑miR451* and ↑miR454*. Current modern diagnostic laboratory method for identification of coronary obstruction is still in its early stage. Nevertheless, current molecular medicine and diagnostic methods are capable to identify if an individual is predisposed to the illness. Platelet miRNA expression profile detection is associated with platelet hyperactivity and may serve as an important marker for prevention of coronary artery disease (CAD) in human overweight/obesity.
Abbreviations
ADP: Adenosine diphosphate
BFGF: Basic Fibroblast Growth Factor
CAD: coronary artery disease
cAMP: cyclic adenosine monophosphate
CCL5: RANTES
CD40L: CD40 Ligand
CHD: Coronary heart disease
CVDs: cardiovascular disorders
CXC: Chemokines
CXCL4: Chemokine ligand 4
DAG: 1,2-diacylglycerol
DGCR 8: di-george syndrome critical region-8
DNA: Deoxyribonucleic Acid
EGF: Epidermal Growth Factor
ENA: 78- epithelial neutrophil activating protein 78
IL-1β: interleukin -1β
IP3: inositol 1,4,5 triphosphate.
miRNA: microRNA
mRNA: messenger RNA
NSTEM: Non Segment Elevation Myocardial Infarction;
ObRb: Leptin receptor
PAI-1: Plasminogen Activator Inhibitor,
PDGF: Platelet-derived growth factor
PF4: platelet factor 4
piRNA: piwi-interacting RNAs
PKC: protein kinase C
PLC: phospholipase C
RISC: RNA In-duced Silencing Complex
siRNA: small interfering RNAs
SNP: single-nucleotide polymorphism
STEM: Segment Elevation Myocardial Infarction
TFPI: tissue factor pathway inhibitor,
TGF-β: Transforming growth factor beta
TRBP: Transactivation Response Binding Protein
UTR: untranslated region
VAMP8: Vesicle-associated Membrane Protein 8
VCAM-1: Vascular Cell Adhesion Molecules-1
VSMC: Vascular Smooth Muscle Cells
vWF: von willbrand factor
WHO: World Health Organization
Competing Interests
The author declares no conflict of interest regarding the article for publication.
Authors' Contributions
Sabariah Md Noor: initiate the conception and technicality; Azrina Azlan, and Loh Su Peng: guide the article publication along with flow of idea and amending the figure respectively; Yakubu Abdulrahman: do the written and revision of the paper.
Acknowledgments
This work was supported by the Geran Putra IPS 957600 (Putra Grant Initiative) from Universiti Putra Malaysia (UPM).
References
-
Purkayastha
S.,
Cai
D.,
Neuroinflammatory basis of metabolic syndrome. Mol Metab.
2013;
2
(4)
:
356-63
.
View Article PubMed Google Scholar -
Heap
G.A.,
van Heel
D.A.,
The genetics of chronic inflammatory diseases. Hum Mol Genet.
2009;
18
:
101-6
.
View Article PubMed Google Scholar -
Obesity Update
O.E.,
Obesity Update 2017. Diabetologe (Heidelb).
2017;
13
(5)
:
331-41
.
View Article Google Scholar -
Mazzoccoli
G.,
Copetti
M.,
Dagostino
M.P.,
Grilli
M.,
Fontana
A.,
Pellegrini
F.,
Epicardial adipose tissue and idiopathic deep venous thrombosis: an association study. Atherosclerosis.
2012;
223
(2)
:
378-83
.
View Article PubMed Google Scholar -
Vayá
A.,
Martínez-Triguero
M.L.,
España
F.,
Todolí
J.A.,
Bonet
E.,
Corella
D.,
The metabolic syndrome and its individual components: its association with venous thromboembolism in a Mediterranean population. Metab Syndr Relat Disord.
2011;
9
(3)
:
197-201
.
View Article PubMed Google Scholar -
Werner
N.,
Nickenig
G.,
From fat fighter to risk factor: the zigzag trek of leptin. Arterioscler Thromb Vasc Biol.
2004;
24
(1)
:
7-9
.
View Article PubMed Google Scholar -
Sader
S.,
Nian
M.,
Liu
P.,
Leptin: a novel link between obesity, diabetes, cardiovascular risk, and ventricular hypertrophy. Circulation.
2003;
108
(6)
:
644-6
.
View Article PubMed Google Scholar -
Yang
R.,
Barouch
L.A.,
Leptin signaling and obesity: cardiovascular consequences. Circ Res.
2007;
101
(6)
:
545-59
.
View Article PubMed Google Scholar -
Wallace
A.M.,
McMahon
A.D.,
Packard
C.J.,
Kelly
A.,
Shepherd
J.,
Gaw
A.,
Plasma leptin and the risk of cardiovascular disease in the west of Scotland coronary prevention study (WOSCOPS). Circulation.
2001;
104
(25)
:
3052-6
.
View Article PubMed Google Scholar -
Nakata
M.,
Yada
T.,
Soejima
N.,
Maruyama
I.,
Leptin promotes aggregation of human platelets via the long form of its receptor. Diabetes.
1999;
48
(2)
:
426-9
.
View Article PubMed Google Scholar -
Konstantinides
S.,
Schäfer
K.,
Koschnick
S.,
Loskutoff
D.J.,
Leptin-dependent platelet aggregation and arterial thrombosis suggests a mechanism for atherothrombotic disease in obesity. J Clin Invest.
2001;
108
(10)
:
1533-40
.
View Article PubMed Google Scholar -
Mitchell
A.J.,
Gray
W.D.,
Hayek
S.S.,
Ko
Y.A.,
Thomas
S.,
Rooney
K.,
Platelets confound the measurement of extracellular miRNA in archived plasma. Sci Rep.
2016;
6
(September)
:
32651
.
View Article PubMed Google Scholar -
Edelstein
L.C.,
Nagalla
S.,
Bray
P.F.,
MicroRNAs in Platelet Production and Activation. MicroRNAs Med..
2013;
2013
:
101-116
.
View Article Google Scholar -
Stern
J.H.,
Rutkowski
J.M.,
Scherer
P.E.,
Adiponectin, Leptin, and Fatty Acids in the Maintenance of Metabolic Homeostasis through Adipose Tissue Crosstalk. Cell Metab.
2016;
23
(5)
:
770-84
.
View Article PubMed Google Scholar -
Jr
M.G.,
Leibel
R.L.,
Seeley
R.J.,
Schwartz
M.W.,
Obesity and Leptin Resistance: Distinguishing Cause from Effect. Trends Endocrinol Metab.
2011;
21
:
643-51
.
View Article Google Scholar -
Mantzoros
C.S.,
Magkos
F.,
Brinkoetter
M.,
Sienkiewicz
E.,
Dardeno
T.A.,
Kim
S.Y.,
Leptin in human physiology and pathophysiology. Am J Physiol Endocrinol Metab.
2011;
301
(4)
:
567-84
.
View Article PubMed Google Scholar -
Elbatarny
H.S.,
Maurice
D.H.,
Leptin-mediated activation of human platelets: involvement of a leptin receptor and phosphodiesterase 3A-containing cellular signaling complex. Am J Physiol Endocrinol Metab.
2005;
289
(4)
:
695-702
.
View Article PubMed Google Scholar -
Bełtowski
J.,
Role of leptin in blood pressure regulation and arterial hypertension. J Hypertens.
2006;
24
(5)
:
789-801
.
View Article PubMed Google Scholar -
Paz-Filho
G.,
Mastronardi
C.,
Franco
C.B.,
Wang
K.B.,
Wong
M.L.,
Licinio
J.,
Leptin: molecular mechanisms, systemic pro-inflammatory effects, and clinical implications. Arq Bras Endocrinol Metabol.
2012;
56
(9)
:
597-607
.
View Article PubMed Google Scholar -
Allman
M.,
Wallace
M.,
Gaskin
L.,
Rivera
C.A.,
Leptin induces an inflammatory phenotype in lean Wistar rats. Mediators Inflamm.
2009;
2009
:
738620
.
View Article PubMed Google Scholar -
Chen
T.H.,
Lee
C.J.,
Wang
J.H.,
Hsu
B.G.,
High Serum Leptin Level is Associated with Peripheral Artery Disease in Geriatric Individuals. Int J Gerontol.
2018;
12
(3)
:
191-5
.
View Article Google Scholar -
Ozata
M.,
Avcu
F.,
Durmus
O.,
Yilmaz
I.,
Ozdemir
I.C.,
Yalcin
A.,
Leptin does not play a major role in platelet aggregation in obesity and leptin deficiency. Obes Res.
2001;
9
(10)
:
627-30
.
View Article PubMed Google Scholar -
Mao
X.,
Ait-Aissa
K.,
Lagrange
J.,
Youcef
G.,
Louis
H.,
Hypertension, hypercoagulability and the metabolic syndrome: a cluster of risk factors for cardiovascular disease. Biomed Mater Eng.
2012;
22
(1-3)
:
35-48
.
View Article PubMed Google Scholar -
Xia
L.,
Zeng
Z.,
Tang
W.H.,
The Role of Platelet Microparticle Associated microRNAs in Cellular Crosstalk. Front. Cardiovasc. Med..
2018;
5
:
29
.
View Article Google Scholar -
Dou
L.,
Zhao
T.,
Wang
L.,
Huang
X.,
Jiao
J.,
Gao
D.,
miR-200s contribute to interleukin-6 (IL-6)-induced insulin resistance in hepatocytes. J Biol Chem.
2013;
288
(31)
:
22596-606
.
View Article PubMed Google Scholar -
Benoit
C.,
Ould-Hamouda
H.,
Crepin
D.,
Gertler
A.,
Amar
L.,
Taouis
M.,
Early leptin blockade predisposes fat-fed rats to overweight and modifies hypothalamic microRNAs. J Endocrinol.
2013;
218
(1)
:
35-47
.
View Article PubMed Google Scholar -
Sharda
A.,
Flaumenhaft
R.,
The life cycle of platelet granules. F1000 Res.
2018;
7
(0)
:
236
.
View Article PubMed Google Scholar -
Bijak
M.,
Saluk
J.,
Ponczek
M.B.,
Nowak
P.,
Wachowicz
B.,
The synthesis of proteins in unnucleated blood platelets. Postepy Hig Med Dosw.
2013;
506
:
672-679
.
PubMed Google Scholar -
Patil
V.S.,
Zhou
R.,
Rana
T.M.,
Gene regulation by non-coding RNAs. Crit Rev Biochem Mol Biol.
2014;
49
(1)
:
16-32
.
View Article PubMed Google Scholar -
Britten
R.J.,
Davidson
E.H.,
Dike
S.,
Gene Regulation for Higher Cells: A Theory. Science.
1969;
165
(3891)
:
349-357
.
View Article Google Scholar -
Ui-Tei
K.,
Naito
Y.,
Nishi
K.,
Juni
A.,
Saigo
K.,
Thermodynamic stability and Watson-Crick base pairing in the seed duplex are major determinants of the efficiency of the siRNA-based off-target effect. Nucleic Acids Res.
2008;
36
(22)
:
7100-9
.
View Article PubMed Google Scholar -
Landry
P.,
Plante
I.,
Ouellet
D.L.,
Perron
M.P.,
Rousseau
G.,
Provost
P.,
Existence of a microRNA pathway in anucleate platelets. Nat Struct Mol Biol.
2009;
16
(9)
:
961-6
.
View Article PubMed Google Scholar -
Small
E.M.,
Olson
E.N.,
Pervasive roles of microRNAs in cardiovascular biology. Nature.
2011;
469
(7330)
:
336-42
.
View Article PubMed Google Scholar -
Bartel
D.P.,
MicroRNAs: target recognition and regulatory functions. Cell.
2009;
136
(2)
:
215-33
.
View Article PubMed Google Scholar -
Nagalla
S.,
Shaw
C.,
Kong
X.,
Kondkar
A.A.,
Edelstein
L.C.,
Ma
L.,
Platelet microRNA-mRNA coexpression profiles correlate with platelet reactivity. Blood.
2011;
117
(19)
:
5189-97
.
View Article PubMed Google Scholar -
Lund
E.,
Güttinger
S.,
Calado
A.,
J.E. Dahlberg,
U. Kutay,
Nuclear export of microRNA precursors. Science.
2004;
303
(5654)
:
95-98
.
-
Zamore
P.D.,
Tuschl
T.,
Sharp
P.A.,
Bartel
D.P.,
Phillip
D.,
Double-Stranded RNA Directs the ATP-Dependent Cleavage of mRNA at 21 to 23 Nucleotide Intervals. Zamore Thomas Tuschl..
2000;
101
:
25-33
.
View Article Google Scholar -
Preall
J.B.,
He
Z.,
Gorra
J.M.,
Sontheimer
E.J.,
Short interfering RNA strand selection is independent of dsRNA processing polarity during RNAi in Drosophila. Curr Biol.
2006;
16
(5)
:
530-5
.
View Article PubMed Google Scholar -
Siomi
H.,
Siomi
M.C.,
On the road to reading the RNA-interference code. Nature.
2009;
457
(7228)
:
396-404
.
View Article PubMed Google Scholar -
Gregory
R.I.,
Chendrimada
T.P.,
Cooch
N.,
Shiekhattar
R.,
Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell.
2005;
123
(4)
:
631-40
.
View Article PubMed Google Scholar -
Xie
H.,
Sun
L.,
Lodish
H.F.,
Targeting microRNAs in obesity. Expert Opin Ther Targets.
2009;
13
(10)
:
1227-38
.
View Article PubMed Google Scholar -
Moszyńska
A.,
Gebert
M.,
Collawn
J.F.,
Bartoszewski
R.,
SNPs in microRNA target sites and their potential role in human disease. Open Biol.
2017;
7
(4)
:
170019
.
View Article PubMed Google Scholar -
Zhang
W.,
Wang
L.,
Zhou
P.,
Song
G.,
Shen
M.,
Gan
S.,
Identification and analysis of genetic variations in pri-miRNAs expressed specifically or at a high level in sheep skeletal muscle. PLoS One.
2015;
10
(2)
:
e0117327
.
View Article PubMed Google Scholar -
Króliczewski
J.,
Sobolewska
A.,
Lejnowski
D.,
Collawn
J.F.,
Bartoszewski
R.,
microRNA single polynucleotide polymorphism influences on microRNA biogenesis and mRNA target specificity. Gene.
2018;
640
:
66-72
.
View Article PubMed Google Scholar -
Schwerk
J.,
Savan
R.,
Translating the Untranslated Region. J Immunol.
2015;
195
(7)
:
2963-71
.
View Article PubMed Google Scholar -
Vaishnavi
V.,
Manikandan
M.,
Munirajan
A.K.,
Mining the 3\textquotesingleUTR of autism-implicated genes for SNPs perturbing microRNA regulation. Genomics Proteomics Bioinformatics.
2014;
12
(2)
:
92-104
.
View Article PubMed Google Scholar -
X.D. Xiong,
M. Cho,
X.P. Cai,
J. Cheng,
X. Jing,
J.M. Cen,
X. Liu,
X.L. Yang,
Y. Suh,
A common variant in pre-miR-146 is associated with coronary artery disease risk and its mature miRNA expression. Mutat Res - Fundam Mol Mech Mutagen. .
2014;
761
:
15-20
.
View Article Google Scholar -
Wang
Y.,
Wang
X.,
Li
Z.,
Chen
L.,
Zhou
L.,
Li
C.,
Two single nucleotide polymorphisms (rs2431697 and rs2910164) of miR-146a are associated with risk of coronary artery disease. Int J Environ Res Public Health.
2017;
14
(5)
:
4-10
.
View Article PubMed Google Scholar -
Liu
X.,
You
L.,
Zhou
R.,
Zhang
J.,
Significant association between functional microRNA polymorphisms and coronary heart disease susceptibility% UNKNOWN UNICODE CHARACTER 0202F (NARROW NO-BREAK SPACE) : a comprehensive meta-analysis involving 16484 subjects. Oncotarget.
2016;
\textbullet\textbullet\textbullet
:
1-11
.
View Article PubMed Google Scholar -
Xie
X.,
Shi
X.,
Xun
X.,
Rao
L.,
Association between microRNA polymorphisms and coronary heart disease : A meta-analysis. Herz.
2017;
42
(6)
:
593-603
.
View Article PubMed Google Scholar -
Bao
M.H.,
Xiao
Y.,
Zhang
Q.S.,
Luo
H.Q.,
Luo
J.,
Zhao
J.,
Meta-analysis of miR-146a polymorphisms association with coronary artery diseases and ischemic stroke. Int J Mol Sci.
2015;
16
(7)
:
14305-17
.
View Article PubMed Google Scholar -
Duan
X.,
Zhan
Q.,
Song
B.,
Zeng
S.,
Zhou
J.,
Long
Y.,
Detection of platelet microRNA expression in patients with diabetes mellitus with or without ischemic stroke. J Diabetes Complications.
2014;
28
(5)
:
705-10
.
View Article PubMed Google Scholar -
Gatsiou
A.,
Boeckel
J.N.,
Randriamboavonjy
V.,
Stellos
K.,
MicroRNAs in platelet biogenesis and function: implications in vascular homeostasis and inflammation. Curr Vasc Pharmacol.
2012;
10
(5)
:
524-31
.
View Article PubMed Google Scholar -
Mitchell
P.S.,
Parkin
R.K.,
Kroh
E.M.,
Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences.
2008;
105
(30)
:
10513-10518
.
View Article Google Scholar -
Laterza
O.F.,
Lim
L.,
Garrett-Engele
P.W.,
Vlasakova
K.,
Muniappa
N.,
Tanaka
W.K.,
Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem.
2009;
55
(11)
:
1977-83
.
View Article PubMed Google Scholar -
Schulte
C.,
Zeller
T.,
microRNA-based diagnostics and therapy in cardiovascular disease-Summing up the facts. Cardiovasc Diagn Ther.
2015;
5
(1)
:
17-36
.
View Article PubMed Google Scholar -
Pordzik
J.,
Pisarz
K.,
De Rosa
S.,
Jones
A.D.,
Eyileten
C.,
Indolfi
C.,
The potential role of platelet-related microRNAs in the development of cardiovascular events in high-risk populations, including diabetic patients: A review. Front Endocrinol (Lausanne).
2018;
9
(MAR)
:
74
.
View Article PubMed Google Scholar -
Ding
X.,
Ding
J.,
Ning
J.,
Yi
F.,
Chen
J.,
Zhao
D.,
Circulating microRNA-122 as a potential biomarker for liver injury. Mol Med Rep.
2012;
5
(6)
:
1428-32
.
View Article PubMed Google Scholar -
Tijsen
A.J.,
Creemers
E.E.,
Moerland
P.D.,
de Windt
L.J.,
van der Wal
A.C.,
Kok
W.E.,
MiR423-5p as a circulating biomarker for heart failure. Circ Res.
2010;
106
(6)
:
1035-9
.
View Article PubMed Google Scholar -
S. Yang,
J. Zhao,
Y. Chen,
M. Lei,
Biomarkers Associated with Ischemic Stroke in Diabetes Mellitus Patients. Cardiovascular toxicology.
2016;
16
(3)
:
213-222
.
-
Santilli
F.,
Vazzana
N.,
Liani
R.,
Guagnano
M.T.,
Dav\`\i
G.,
Platelet activation in obesity and metabolic syndrome. Obes Rev.
2012;
13
(1)
:
27-42
.
View Article PubMed Google Scholar -
F. Santilli,
N. Vazzana,
R. Liani,
M. Guagnano,
G. Davì,
Platelet activation in obesity and metabolic syndrome. . doi:. Obes Rev..
2012;
13
(1)
:
27-42
.
View Article Google Scholar -
Yang
W.J.,
Yang
D.D.,
Na
S.,
Sandusky
G.E.,
Zhang
Q.,
Zhao
G.,
Dicer Is Required for Embryonic Angiogenesis during Mouse Development. Journal of Biological Chemistry.
2005;
280
(1)
:
9330-9335
.
View Article Google Scholar -
Duan
R.,
Pak
C.,
Jin
P.,
Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Human molecular genetics.
2007;
16
(9)
:
1124-1131
.
View Article Google Scholar -
Albinsson
S.,
Suarez
Y.,
Skoura
A.,
Offermanns
S.,
Joseph
M.,
Sessa
W.C.,
differentiation and function. 2011;
30
(6)
:
1118-1126
.
View Article Google Scholar -
Altuvia
Y.,
Landgraf
P.,
Lithwick
G.,
Elefant
N.,
Pfeffer
S.,
Aravin
A.,
Clustering and conservation patterns of human microRNAs. Nucleic Acids Res.
2005;
33
(8)
:
2697-706
.
View Article PubMed Google Scholar -
Zhu
N.,
Zhang
D.,
Chen
S.,
Liu
X.,
Lin
L.,
Huang
X.,
Endothelial enriched microRNAs regulate angiotensin II-induced endothelial inflammation and migration. Atherosclerosis.
2011;
215
(2)
:
286-93
.
View Article PubMed Google Scholar -
Hartmann
D.,
Thum
T.,
MicroRNAs and vascular (dys)function. Vascul Pharmacol.
2011;
55
(4)
:
92-105
.
View Article PubMed Google Scholar -
Lovren
F.,
Pan
Y.,
Quan
A.,
Singh
K.K.,
Shukla
P.C.,
Gupta
N.,
MicroRNA-145 targeted therapy reduces atherosclerosis. Circulation.
2012;
126
(11)
:
81-90
.
View Article PubMed Google Scholar -
Lu
M.,
Zhang
Q.,
Deng
M.,
Miao
J.,
Guo
Y.,
Gao
W.,
An analysis of human microRNA and disease associations. PLoS One.
2008;
3
(10)
:
e3420
.
View Article PubMed Google Scholar -
Nieuwdorp
M.,
Stroes
E.S.G.,
Meijers
J.C.M.,
Büller
H.,
Hypercoagulability in the metabolic syndrome. Curr Opin Pharmacol. .
2005;
5
(2 Spec. Iss)
:
155-159
.
View Article Google Scholar -
Zimmerman
G.a.,
A.S. Weyrich,
Pathways to Altered Phenotype and Function. Arterioscler Thromb.
2008;
28
(3)
.
View Article Google Scholar -
Schulte
C.,
Molz
S.,
Appelbaum
S.,
Karakas
M.,
Ojeda
F.,
Lau
D.M.,
MiRNA-197 and miRNA-223 predict cardiovascular death in a cohort of patients with symptomatic coronary artery disease. PLoS One.
2015;
10
(12)
:
e0145930
.
View Article PubMed Google Scholar -
Goren
Y.,
Meiri
E.,
Hogan
C.,
Mitchell
H.,
Lebanony
D.,
Salman
N.,
Relation of reduced expression of MiR-150 in platelets to atrial fibrillation in patients with chronic systolic heart failure. Am J Cardiol.
2014;
113
(6)
:
976-81
.
View Article PubMed Google Scholar -
JA
Ward,
N
Esa,
R
Pidikiti,
JE
Freedman,
JF
Keaney,
K
Tanriverdi,
O
Vitseva,
V
Ambros,
R
Lee,
DD
McManus,
Circulating cell and plasma microRNA profiles differ between non-ST-segment and ST-segment-elevation myocardial infarction. Family medicine & medical science research.
2014;
2
(2)
:
108
.
View Article Google Scholar -
Hollopeter
G.,
Jantzen
H.M.,
Vincent
D.,
Li
G.,
England
L.,
Ramakrishnan
V.,
Identification of the platelet ADP receptor targeted by antithrombotic drugs. Nature.
2001;
409
(6817)
:
202-7
.
View Article PubMed Google Scholar -
Cavarretta
E.,
Chiariello
G.A.,
Condorelli
G.,
Platelets, endothelium, and circulating microRNA-126 as a prognostic biomarker in cardiovascular diseases: per aspirin ad astra. Eur Heart J.
2013;
34
(44)
:
3400-2
.
View Article PubMed Google Scholar -
Somanath
P.R.,
Podrez
E.A.,
Chen
J.,
Ma
Y.I.,
Antoch
M.,
Byzova
T.V.,
Deficiency in core circadian protein Bmal1 is associated with a prothrombotic and vascular phenotype. Journal of cellular physiology.
2011;
226
(1)
:
132-40
.
View Article Google Scholar -
Shan
S.W.,
Lee
D.Y.,
Deng
Z.,
Shatseva
T.,
Jeyapalan
Z.,
Du
W.W.,
MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat Cell Biol.
2009;
11
(8)
:
1031-8
.
View Article PubMed Google Scholar -
Fichtlscherer
S.,
De Rosa
S.,
Fox
H.,
Schwietz
T.,
Fischer
A.,
Liebetrau
C.,
Circulating microRNAs in patients with coronary artery disease. Circ Res.
2010;
107
(5)
:
677-84
.
View Article PubMed Google Scholar -
Peng
H.,
Zhong
M.,
Zhao
W.,
Wang
C.,
Zhang
J.,
Liu
X.,
Urinary miR-29 correlates with albuminuria and carotid intima-media thickness in type 2 diabetes patients. PLoS One.
2013;
8
(12)
:
e82607
.
View Article PubMed Google Scholar -
van Rooij
E.,
Sutherland
L.B.,
Thatcher
J.E.,
DiMaio
J.M.,
Naseem
R.H.,
Marshall
W.S.,
Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA.
2008;
105
(35)
:
13027-32
.
View Article PubMed Google Scholar -
Shiffman
D.,
Rowland
C.M.,
Louie
J.Z.,
Luke
M.M.,
Bare
L.A.,
Bolonick
J.I.,
Gene variants of VAMP8 and HNRPUL1 are associated with early-onset myocardial infarction. Arterioscler Thromb Vasc Biol.
2006;
26
(7)
:
1613-8
.
View Article PubMed Google Scholar -
Ezzell
R.M.,
Kenney
D.M.,
Egan
S.,
Stossel
T.P.,
Hartwig
J.H.,
Localization of the domain of actin-binding protein that binds to membrane glycoprotein Ib and actin in human platelets. J Biol Chem.
1988;
263
(26)
:
13303-9
.
PubMed Google Scholar -
Elgheznawy
A.,
Fleming
I.,
Platelet-enriched microRNAs and cardiovascular homeostasis. Antioxid Redox Signal.
2017;
2017
(9)
.
View Article Google Scholar -
Harris
T.A.,
Yamakuchi
M.,
Ferlito
M.,
Mendell
J.T.,
Lowenstein
C.J.,
MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA.
2008;
105
(5)
:
1516-21
.
View Article PubMed Google Scholar -
Gambaryan
S.,
Kobsar
A.,
Rukoyatkina
N.,
Herterich
S.,
Geiger
J.,
Smolenski
A.,
Thrombin and collagen induce a feedback inhibitory signaling pathway in platelets involving dissociation of the catalytic subunit of protein kinase A from an NFkappaB-IkappaB complex. J Biol Chem.
2010;
285
(24)
:
18352-63
.
View Article PubMed Google Scholar -
Choi
Y.C.,
Yoon
S.,
Jeong
Y.,
Yoon
J.,
Baek
K.,
Regulation of vascular endothelial growth factor signaling by miR-200b. Mol Cells.
2011;
32
(1)
:
77-82
.
View Article PubMed Google Scholar -
Fasanaro
P.,
D'Alessandra
Y.,
Di Stefano
V.,
Melchionna
R.,
Romani
S.,
Pompilio
G.,
MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem.
2008;
283
(23)
:
15878-83
.
View Article PubMed Google Scholar -
Guduric-Fuchs
J.,
O'Connor
A.,
Cullen
A.,
Harwood
L.,
Medina
R.J.,
O'Neill
C.L.,
Deep sequencing reveals predominant expression of miR-21 amongst the small non-coding RNAs in retinal microvascular endothelial cells. J Cell Biochem.
2012;
113
(6)
:
2098-111
.
View Article PubMed Google Scholar -
Fleissner
F.,
Jazbutyte
V.,
Fiedler
J.,
Gupta
S.K.,
Yin
X.,
Xu
Q.,
Short communication: asymmetric dimethylarginine impairs angiogenic progenitor cell function in patients with coronary artery disease through a microRNA-21-dependent mechanism. Circ Res.
2010;
107
(1)
:
138-43
.
View Article PubMed Google Scholar -
Plé
H.,
Landry
P.,
Benham
A.,
Coarfa
C.,
Gunaratne
P.H.,
Provost
P.,
The repertoire and features of human platelet microRNAs. PLoS One.
2012;
7
(12)
:
e50746
.
View Article PubMed Google Scholar
Comments
Downloads
Article Details
Volume & Issue : Vol 6 No 6 (2019)
Page No.: 3248-3261
Published on: 2019-07-03
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 6089 times
- Download PDF downloaded - 1340 times
- View Article downloaded - 0 times
 Biomedpress
Biomedpress