Abstract
Introduction: Urinary tract infections (UTIs) are an increasing public health problem caused by a range of uropathogens. UTI is a major health concern among tribal women due to lack of adequate health facilities at regional public health centers and district hospitals. The objective of the study was to investigate the prevalence and antibiotic susceptibility of bacteria isolated from urine samples of tribal women suffering from UTI. Urine samples were collected from 550 tribal women who visited the Outpatient Department (OPD) of the District Hospital of Anuppur, Madhya Pradesh, India.
Methods: The samples were cultured on different selective and differential media and identified by colony morphology, Gram staining and biochemical tests.
Results: Out of 550 urine samples, 360 (65.45%) were culture-positive and 190 (34.55%) were culture-negative. Klebsiella pneumoniae, Proteus mirabilis, Escherichia coli & Pseudomonas aeruginosa have been identified as major uropathogens. K. pneumoniae is the most prevalent bacteria followed by P. mirabilis, E. coli and P. aeruginosa. The drug sensitivity analysis of bacterial uropathogens was performed against different antibiotics. The antibiotic susceptibility profiling showed that most of the isolates of K. pneumoniae, P. mirabilis and E. coli were resistant against penicillin (86.67-100%) and rifampicin (72.73-97.67%).
Conclusions: Gentamycin, kanamycin and streptomycin were the most active antibiotics against the isolates of P. aeruginosa (76, 80 and 76%), P. mirabilis (76.62, 66.23 and 75.32%), K. pneumoniae (55.81, 20.93 and 44.19%) and E. coli (50.48, 37.14 and 62.86%), respectively, and may be the drug of choice for treatment of UTI in this region at present.
Introduction
Urinary Tract Infections (UTIs) are the most common infections, affecting almost 50% of the human population, particularly women, at least once in their lifetime1. It is one of the major causes of morbidities and discomfort among women, especially in developing countries. UTI is the third most common infection in India, which affects women of all age groups2. Acute and uncomplicated UTIs are most commonly found in women and it has been estimated that more than 60% of women have UTI at least once in their lifetime 1. It has also been reported that the rate of UTI is 10.57% higher in sexually active females and teenage girls. Although different microbes such as Acinetobacter baumannii, Citrobacter freundii, Enterobacter aerogenes, Enterobacter cloacae, Enterococcus faecalis, Enterococcus faecium, Escherichia coli, Klebsiella oxytoca, Klebsiella pneumoniae, Morganella morganii, Proteus mirabilis, Proteus vulgaris, Providencia stuartii, Pseudomonas aeruginosa, Staphylococcus saprophyticus, Streptococcus agalactiae and yeast (Candida albicans) have been reported in UTIs, but bacterial isolates are most prevalent3. The most common bacteria involved in UTIs are E. coli, K. pneumoniae and Staphylococcus aureus4.
UTI may be treated easily on the identification of causative bacteria followed by the analysis of antibiotic susceptibility pattern and prescription of suitable antibiotics5. The non-judicious use of antibiotics without antibiotic susceptibility tests, self-medication, overdose and misuse of antibiotics has led to the development of antibiotic resistance, resulting in treatment failure6. According to the World Health Organization report of 2014, increasing antimicrobial resistance is a global threat for public health, especially in developing countries, such that the problem must be dealt with high priority7.
The schedule tribes (STs) are the weakest section of the Indian population from the economic and educational perspectives. In tribal societies, the women are the main sufferers. The tribal women carry the major burden of disease due to multiple factors such as poverty, illiteracy and lack of awareness, malnutrition, unhygienic habitat and superstitions. Due to poor personal hygiene and sanitation, UTIs have become widely prevalent among the tribal women. UTI not only affects the quality of life of the women but is a social stigma in less educated tribal societies.
The study herein aimed to investigate the bacteria associated with UTI and their antibiotic susceptibility patterns among tribal women in District Anuppur, Madhya Pradesh, India. The study will help in formulating guidelines for establishing proper empirical therapies for UTIs in this region.
Materials - Methods
Collection of Urine Specimens
The urine samples were collected from women having symptoms of UTI, who visited the Out Patient Department (OPD) of the District Hospital, Anuppur, Madhya Pradesh, India. To avoid contamination, midstream urine was collected from suspected UTI patients in sterile tubes and stored in the refrigerator (4 0C) unit until use.
Quantitative analysis
Quantitative analysis of microbes in the urine sample was determined as cfu/ml (colony forming units per ml) by plate count method (spread plate technique) on nutrient agar. Urine samples with significant bacterial growth (≥ 105 cfu/ml) were considered to be culture-positive and processed further. Samples with no growth or growth ≤ 105 Cfu/ml were considered culture-negative8.
Isolation and identification of bacteria
Bacteria were isolated & identified using conventional microbiological methods. Selective and differential media (such as MacConkey, Hi Chrome UTI Agar and Cetrimide Agar) were purchased from Hi-Media (Mumbai, India) and were used for identification of bacteria. Briefly, 0.01 ml of positive urine sample was streaked aseptically on various selective and differential media. The plates were incubated aerobically at 37°C for 24 hours. The uropathogens were tentatively identified based on their growth in selective and differential media, color generation, as well as colony morphology. The individual colonies from the selective and differential media were further subcultured to isolate the single pure culture of the selected bacteria. The identity of the isolated pure culture was further established by colony morphology, Gram staining and biochemical tests9.
Antimicrobial sensitivity test
The antimicrobial susceptibility of the isolated and identified bacteria against the commonly prescribed antibiotics was carried out by the Kirby-Bauer disk diffusion method. Sterile Mueller-Hinton agar (MHA) plates were prepared for this test. Using sterile cotton swabs, the test organisms were inoculated on MHA plates for preparing bacterial lawn. The cultures were allowed to dry on the plate for 5-10 minutes at room temperature in laminar airflow. Discs of selected antibiotics were placed on the surface of the agar medium. The discs were gently pressed with the help of sterile forceps for better contact and effective diffusion of the antibiotics into the medium. The plates were incubated in an inverted position for 16-18 hours at 370C and the zone of inhibition was measured for estimation of antibiotic sensitivity10.
Drug sensitivity profiling was assessed for each antibiotic as ‘Sensitive’, ‘Resistant’ or ‘Intermediate’, as per the measured diameter of growth of inhibition zone in millimeters (mm) as shown in Table 1, and as per previously established methods11.
Antibiotics used for the study
The commonly prescribed antibiotics for the treatments of UTI were selected for susceptibility testing. The discs of gentamicin (GEN, 10 µg), ampicillin (AMP, 10 µg), penicillin-G (P, 10 µg), kanamycin (K, 30 µg), streptomycin (S, 10 µg), norfloxacin (NX, 10 µg), nalidixic acid (NA, 30 µg), amoxicillin (AMC, 30 µg), rifampicin (RIF, 5 µg), chloramphenicol (C, 30 µg), co-trimoxazole (COT, 25 µg), tetracycline (TE, 30 µg) and ciprofloxacin (CIP, 5 µg) were procured from Hi Media.
| Antibiotics | Disc concn | Diameter of growth inhibition zone (mm ) | ||
| Sensitive | Intermediate | Resistant | ||
| Ampicillin (AMP) | 10 mcg | 14 or more | 12 – 13 | 11 or less |
| Amoxicillin (AMC) | 30 mcg | 18 or more | 14 – 17 | 13 or less |
| Chloramphenicol (C) | 30 mcg | 18 or more | 13 – 14 | 12 or less |
| Co–trimoxazole (COT) | 25 mcg | 16 or more | 11 – 15 | 10 or less |
| Ciprofloxacin (CIP) | 5 mcg | 18 or more | 16 – 20 | 15 or less |
| Gentamicin (GEN) | 10 mcg | 15 or more | 13 – 14 | 12 or less |
| Kanamycin (K) | 30 mcg | 18 or more | 14 – 17 | 13 or less |
| Norfloxacin (NX) | 10 mcg | 17 or more | 13 – 16 | 12 or less |
| Nalidixic acid (NA) | 30 mcg | 19 or more | 14 – 18 | 13 or less |
| Penicillin-G (P) | 10 mcg | 29 or more | 21 – 28 | 20 or less |
| Rifampicin (RIF) | 5 mcg | 20 or more | 17 – 19 | 16 or less |
| Streptomycin (S) | 10 mcg | 15 or more | 12 – 14 | 11 or less |
| Tetracycline (TE) | 30 mcg | 19 or more | 15 – 18 | 14 or less |
Results
Quantitative analysis of urine culture
Out of a total of 550 urine samples collected for analysis, 360 (65.45%) were culture-positive with bacteriuria ≥ 105 cfu/ml and 190 (34.55%) were culture-negative with no growth or growth ≤ 105 cfu/ml.
Isolation and identification of uropathogens in selective and differential media
The bacteria from culture-positive samples were tentatively identified based on specific color produced on various selective and differential media. On MacConkey agar media, E. coli and K. pneumoniae showed red/pink colonies while P. mirabilis and P. aeruginosa were observed as colorless. On Hi Chrome UTI media, E. coli produced magenta color, K. pneumoniae produced blue color, while P. mirabilis and P. aeruginosa produced light brown and colorless colonies, respectively. The Citramide agar was used for the selective culture of P. aeruginosa which showed yellowish colored colonies. Single colonies were further isolated and plated on the fresh selective and differential media, with Hi chrome UTI agar and Cetrimide agar being the most accepted conventional media with specific identifying color characteristics, for further confirmation of different uropathogens.
Identification of bacteria by Gram staining and biochemical tests
The presumptively identified isolated bacteria in culture were further characterized by Gram staining, microscopic examination of colony, and specific biochemical tests viz. catalase, IMViC (Indole, Methyl red, Voges-Proskauer, and Citrate). The results are summarized in Table 2.
| Gram Stain | Shape | Colour of colony in various selective and differential media | Biochemical Tests | ||||||||
| S.No. | Bacteria | MacConkey | Hi Chrome UTI | Cetramide | V | M | Cat | I | C | ||
| 1 | E. coli | (-) | Rod | Red/Pink | Magneta | No growth | (-) | (+) | (+) | (+) | (-) |
| 2 | K. pneumoniae | (-) | Rod | Pink (mucoid) | Blue | No growth | (+) | (-) | (+) | (-) | (+) |
| 3 | P. mirabilis | (-) | Rod | Colourless | Light brown | No growth | (-) | (+) | (+) | (-) | (+) |
| 4 | P. pseudomonas | (-) | Rod | Colourless | Colourless | Yellow | (-) | (-) | (+) | (-) | (+) |
Prevalence of different bacterial isolates in urine samples
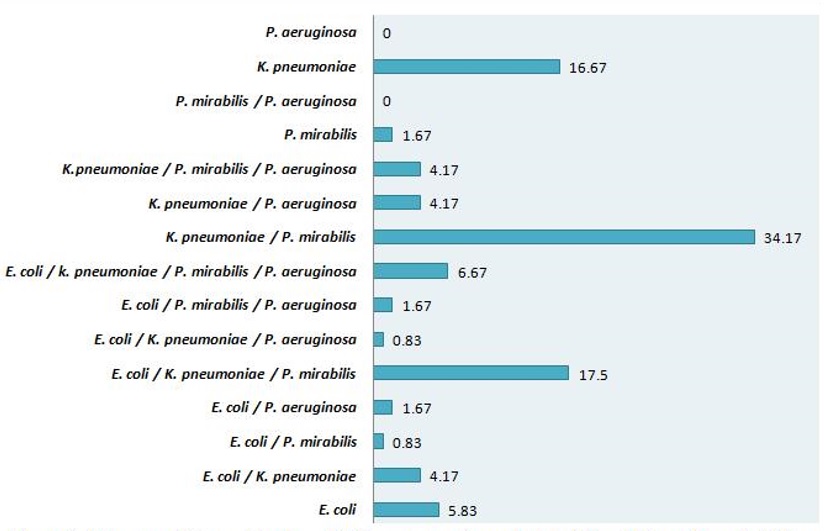
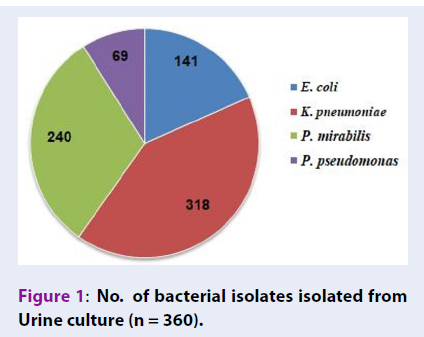
The uropathogens were isolated and identified from 360 culture-positive urine samples. K. pneumoniae (n = 318) was the most prevalent isolate followed by P. mirabilis (n = 240), E. coli (n = 141) and P. aeruginosa (n = 69) (Figure 1). The results also showed the presence of polymicrobial culture growth, i.e. the various combinations of bacteria belonging to different genera, in urine culture. The combination of K. pneumonia with P. mirabilis was most frequently isolated (34.17%), followed by E. coli/K. pneumoniae/P. mirabilis combination (17.50%) (Figure 2).
Antimicrobial sensitivity profiling
Antibiotic sensitivity profile of E. coli
From the total isolates of E. coli, 63.81% were sensitive to DNA replication inhibitor antibiotics norfloxacin, 62.86% were sensitive to streptomycin, and 50.48% were sensitive to gentamicin. E. coli showed the highest resistance against RNA synthesis inhibitor rifampicin (97.67%), followed by penicillin (86.67%), co-trimoxazole (60.47%) and nalidixic acid (60.47%). The percentage of isolates with intermediate sensitivity were low for most of the antibiotics except for tetracycline (44.19%) (Figure 3).
Antibiotic sensitivity profile of K. pneumoniae
From the total isolates of K. pneumoniae, 55.81% were sensitive to protein synthesis inhibitor antibiotic gentamicin, 48.84% to chloramphenicol, and 46.51% to ciprofloxacin. K. pneumoniae showed the highest resistance against β-lactam antibiotic penicillin (100%), followed by rifampicin (97.67%) and ampicillin (72.09%). Most of the isolates were either sensitive or resistant to the tested antibiotics. The number of isolates with intermediate sensitivity was very low for most of the antibiotics except for tetracycline (44.19%) (Figure 4).
Antibiotic sensitivity profile of P. mirabilis
From the total isolates of P. mirabilis, 76.62% were sensitive to protein synthesis inhibitor antibiotic gentamycin and 75.32% to streptomycin. Also, > 60% isolates of P. mirabilis were sensitive to amoxicillin, kanamycin, ampicillin and norfloxacin. P. mirabilis showed the highest resistance against β-lactam antibiotic penicillin (100%), followed by rifampicin (72.73%) and nalidixic acid (55.84%). The intermediate sensitivity of isolates was low for most of the antibiotics except for co-trimoxazole (50.65%) (Figure 5).
Antibiotic sensitivity profile of P. aeruginosa
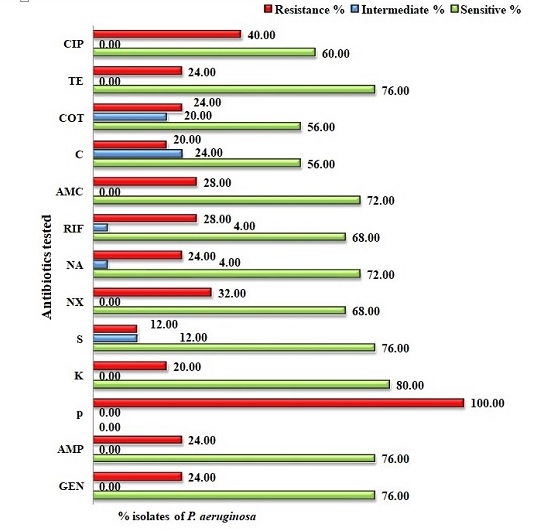
P. aeruginosa isolates showed 76% sensitivity to the protein synthesis inhibitor gentamycin, and > 70% sensitivity to ampicillin, tetracycline, streptomycin, amoxicillin and nalidixic acid. P. aeruginosa showed the highest resistance against β-lactam antibiotic penicillin (100%) (Figure 6).
Discussion
Gram-negative bacteria are the chiefly isolated causative organisms from UTI patients. In our study, K. pneumoniae was the most prevalent (88.33 %) bacteria alone and in combination with other bacterial isolates; it was responsible for infections in tribal women of this demography, followed by P. mirabilis (66.67%). Although E. coli has been reported as the major causative organism in UTIs accounting for 75% to 90% of UTI isolates, K. pneumoniae (65%), P. aeruginosa (50%), Staphylococcus (20%), P. mirabilis (5%) and Candida Spp. (5%) have also been prevalent UTI isolates among the Paliyar tribal group of India12.
Chopra et al., in 2008, reported that the most frequently isolated UTI pathogens in decreasing order were Klebsiella, E. coli, Pseudomonas and Enterobacter 13. The prevalence of K. pneumonia has also been reported to increase in UTIs mainly due to increased antibiotic-resistant phenotypes14. Therefore, our findings of Klebsiella being the most prevalent among the isolates is in accordance with some of the earlier reports.
The updated knowledge of prevailing bacterial uropathogens and resistivity for the various antibiotics in use is of prime importance for the proper use of antimicrobial drugs and empirical treatment of UTIs. Poor clinical practices as well as non-judicious use of antibiotics has led to the development of multi-drug resistant (MDR) strains15. Hence, the evaluation of antimicrobial sensitivity of the isolated pathogens is of paramount importance for effective treatment of UTI.
In the present study, we observed that the broad spectrum β-lactam aminopenicillins, ampicillin and amoxicillin, showed better efficacy as compared to the narrow spectrum β-lactam antibiotic, penicillin. All the major clinical isolates showed a high degree of resistance against penicillin, e.g. E. coli (86.67%), K. pneumoniae (100%), P. mirabilis (100%), P. aeruginosa (100%). Ampicillin and amoxicillin showed moderate to good activity by inhibiting isolates of E. coli (48.57 and 44.19%), P. mirabilis (63.64 and 67.53%) and P. aeruginosa (76 and 72%), respectively. Amoxicillin was active against 44.19% isolates of K. pneumoniae but ampicillin was only active against 25.58% of isolates. Possibly, the high resistance against penicillin may be due to it being a narrow spectrum antibiotic as well as its indiscriminate usage over the time in this region. An important mechanism of antibiotic resistance among uropathogens is through extended spectrum beta-lactamase (ESBL) production. ESBL hydrolyzes oxyimino-β-lactams such as cephalosporin and monobactum16. E. coli is one of the main ESBL-producing pathogens. ESBL production is transmitted from one bacterium to another through plasmids. These plasmids can carry multiple drug-resistant genes against aminoglycosides, quinolones and sulfamethoxazole at the same time17. ESBL-producing E. coli (ESBL-EC) and ESBL-producing K. pneumoniae (ESBL-KP) are resistant to penicillins, cephalosporins and monobactams. The ESBL producers can also develop co-resistance to other classes of antimicrobial agents, such as fluoroquinolones, co-trimoxazole and aminoglycosides, which are frequently used for UTI treatment18. Penicillin being highly prone to degradation by β-lactamase may be a strong reason for all these bacteria being highly resistant to it. There have been reports of ESBLs being capable of conferring bacterial resistance to the penicillins, first-, second- and third-generation cephalosporins, and aztreonam via hydrolysis of these antibiotics19.
Since β-lactam antibiotics, including penicillins, have been the choice of physicians for treating UTI infections. Prolonged and increased resistance against β-lactam antibiotics can be explained by the production of SHV-1 (β-lactamase encoded on the chromosome or a transferable plasmid) 20. Ampicillin and amoxicillin being of broad-spectrum nature show better activity compared to penicillin.
The protein synthesis inhibitor aminoglycoside, which produces its antibacterial action by binding to the 30S ribosomal subunit of bacteria, was among the most active antibiotics in this study. Gentamycin, kanamycin and streptomycin were active against the isolates of P. aeruginosa (76, 80 and 76%), P. mirabilis (76.62, 66.23 and 75.32%), K. pneumoniae (55.81, 20.93 and 44.19%), and E. coli (50.48, 37.14 and 62.86%), respectively. However, another 30S ribosomal subunit binding protein synthesis inhibitor, tetracycline, showed good to moderate activity against the bacterial isolates of P. aeruginosa (76%), K. pneumoniae and E. coli (44.19%), and P. mirabilis (37.66%). Chloramphenicol which inhibits bacterial protein synthesis by binding to 50S ribosomal subunit showed moderate to low activity against the isolates of P. aeruginosa (56%), K. pneumoniae and E. coli (48.84%), and P. mirabilis (33.77%). The study indicated that aminoglycosides were more active as compared to tetracycline and chloramphenicol.
Quinolones are a recent class of antibiotics introduced for management of UTIs. Out of the three quinolones used as antibiotics in the present study, norfloxacin showed the highest activity against P. aeruginosa (68%), followed by E. coli (63.81%), P. mirabilis (61.04%) and K. pneumoniae (46.51%). Ciprofloxacin was moderately active in P. aeruginosa (60%), followed by P. mirabilis (51.95%), and E. coli and K. pneumoniae (46.51%). Nalidixic acid being the oldest quinolone showed the highest activity against P. aeruginosa (72%), followed by P. mirabilis (36.36%), but showed very poor activity in E. coli and K. pneumoniae (11.63%). Although quinolones are among the most effective drugs in treating UTI, studies have also revealed an increasing resistance to these drugs. Resistance to fluoroquinolones increased from 2.9% in 2000 to 11.3% in 2002 21. The results of other studies have also revealed the resistance of E. coli to ciprofloxacin, the most effective quinolone for treating UTI, and that it increased from 46.6 to 59.4% during the years 1998-200022.
The major finding of our study is the high resistance against rifampicin, a newer generation antibiotic. Isolates of E. coli and K. pneumoniae showed 97.67% resistance and isolates of P. mirabilis showed 72.73% resistance. The antimetabolite sulfa drug co-trimoxazole showed moderate activity in P. aeruginosa (56%), low activity in E. coli and K. pneumonia (30.23 %), and very low activity in P. mirabilis (12.99%).
Resistance to two or more antibiotics has led to the development of MDR strains of uropathogens, rendering treatment difficult. Studies conducted in West Bengal and around other different parts of India have indicated that Klebsiella spp. is the second most resistant uropathogen after E. coli, against various classes of anti-microbial agents23. K. pneumoniae, E. coli and P. mirabilis showed high rates of resistance to ampicillin, streptomycin, amoxicillin, tetracycline, sulfonamide, co-trimoxazole, and carbenicillin-trimethoprim24. In our present study, K. pneumoniae, E. coli, and P. mirabilis showed moderate to high resistance against different classes of antibiotics. The resistance range in K. pneumoniae against the tested antibiotics was from 25.58% (in the case of co-trimoxazole) to 100% (in the case of penicillin); the rest of the antibiotics showed resistance above 50%. Similarly, E. coli and P. mirabilis showed moderate to high resistance to most of the antibiotics. The high MDR in the present study might be due to widespread misuse of antibiotics, inappropriate prescription of drugs, and lack of knowledge about drug resistance in the study area.
Conclusion
Assessment of prevalence and identification of the major uropathogens is of paramount importance for the remotely located tribal districts of India. The high burden of UTIs on tribal women of District Anuppur, Madhya Pradesh, India may be due to their poor hygiene, lack of education and reduced accessibility to a better health facility. Our study identified that penicillin and rifampicin antibiotics should not be prescribed for the management of UTI in this region. To reduce non-judicious use and risk of resistance development, antibiotic sensitivity must be made compulsory for the management of UTI.
Abbreviations
Cfu: Colony Forming Units
MDR: Multi Drug Resistance
UTIs: Urniary Tract Infections
Acknowledgments
The authors are thankful to Indira Gandhi National Tribal University for providing lab facility and District, Hospital Anuppur Madhya Pradesh for providing patient samples.
Author’s contributions
Dr. Poonam Sharma conceptualized, designed, coordinated and drafted the study. Mr. Aashish Kumar Netam performed experiments related to urine culture, isolation and staining of bacteria and antibiotic sensitivity. Dr. Rambir Singh analyzed and interpreted the data and wrote the manuscript. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
Data and materials used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The study was conducted on urine samples of patients who visited the OPD of district hospital for treatment of urinary tract infections. Since urine sample is medical waste material given by the patients, voluntarily, without any invasive sample collection methods, hence Institutional Ethical Committee was not approached for approval of the study protocol. The samples were analyzed anonymously and consent from the patients was not necessary.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
-
Foxman B. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. American Journal of Medicine.
2002;
113
(1)
:
5-13
.
View Article Google Scholar -
Prakash D, Saxena RS. Prevalence and antimicrobial susceptibility pattern of Escherichia coli in hospital acquired and community acquired patients related to urinary tract infection in India. Journal of Applied Pharmaceutical Science.
2013;
3
:
124-132
.
View Article Google Scholar -
Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol.
2015;
13
(5)
:
269-284
.
View Article Google Scholar -
Kumar MS, Maity C, Kumar HS, PT, Kumar KP, Chandra MK. Studies on Drug Sensitivity and Bacterial Prevalence of UTI in Tribal Population of Paschim Medinipur, West Bengal, India. Jundishapur J Microbiol.
2012;
6
:
42-46
.
View Article Google Scholar -
Farrell DJ, Morrissey I, De Rubeis D, Robbins M, Felmingham D. A UK multicentre study of the antimicrobial susceptibility of bacterial pathogens causing urinary tract infection. The Journal of Infection.
2003;
46
(2)
:
94-100
.
View Article PubMed Google Scholar -
Zone CP, Guide S. Antimicrobial resistance and urinary tract infections in the community. Signs.
2017;
6
:
3531-3593
.
-
World Health. "Antimicrobial Resistance: Global Report on Surveillance." World Health Organization, 2014. WH. Antimicrobial resistance: global report on surveillance [Internet]. World Health Organization.
2014
.
-
Piranfar V, Mirnejad R, Erfani M. Incidence and Antibiotic Susceptibility Pattern of Most Common Bacterial Pathogen Causing Urinary Tract Infection (UTI) in Tehran, IRAN, 2012-2013. Int J Enteric Pathogen.
2013;
2
:
e15490
.
View Article Google Scholar -
Tajbakhsh E, Tajbakhsh S, Khamesipour F. Isolation and Molecular Detection of Gram Negative Bacteria Causing Urinary Tract Infection in Patients Referred to Shahrekord Hospitals, Iran. Iranian Red Crescent Medical Journal.
2015;
1
:
e24779
.
View Article Google Scholar -
Singh R, Chandra R, Bose M, Luthra PM. Antibacterial activity of Curcuma longa rhizome extract on pathogenic bacteria. Current Sciences.
2002;
83
:
737-740
.
View Article Google Scholar -
Ouno GA, Korir SC, Cheruiyot JC, et al. Isolation, Identification and Characterization of Urinary Tract Infectious Bacteria and the Effect of Different Antibiotics. Journal of Natural Sciences Researches.
2013;
3
:
150-159
.
-
Prabhu V, Pandian S. Study on the prevalence of urinary tract infection among the paliyar Indian tribe. International Journal of Current Research.
2012;
4
:
44-48
.
-
Chopra I, Schofield C, Everett M, et al. Treatment of health-care-associated infections caused by Gram-negative bacteria: a consensus statement. Lancet Infect Dis.
2008;
8
:
133-139
.
View Article Google Scholar -
Clegg S, Murphy CN. Epidemiology and Virulence of Klebsiella pneumoniae. In: Urinary Tract Infections [Internet]. Washington, DC, USA: ASM Press; [cited 2020 Jun 1].
2016 ;
:
435-457
.
View Article Google Scholar -
Ventola CL. The Antibiotic Resistance Crisis. P T.
2015;
40
(04)
:
277-283
.
View Article Google Scholar -
Mukherjee M, Basu S, Mukherjee SK, Majumder M. Multidrug-Resistance and Extended Spectrum Beta-Lactamase Production in UropathogenicE. coli which were Isolated from Hospitalized Patients in Kolkata, India. J Clin Diagn Res.
2013;
7
(3)
:
449-453
.
View Article PubMed Google Scholar -
Guangying Y, Xueyin Z, Anrong L. Development of a novel test identifying extended-spectrum beta lactamase producing strains relative to resistant plasmids. Chin J Microecol.
2002;
14
:
89-90
.
-
Paterson DL, Bonomo RA. Extended-Spectrum β-Lactamases: a Clinical Update. Clin Microbiol Reviews.
2005;
1
(4)
:
657-686
.
View Article PubMed Google Scholar -
Rawat D, Nair D. Extended-spectrum β-lactamases in Gram Negative Bacteria. Journal of Global Infectious Disesases.
2010;
2
(3)
:
263-274
.
View Article PubMed Google Scholar -
Nugent ME, Hedges RW. The nature of the genetic determinant for the SHV-1 beta-lactamase. Mol Gen Genet.
1979;
175
(3)
:
239-243
.
View Article PubMed Google Scholar -
Kurutepe S, Surucuoglu S, Sezgin C, Gazi H, Gulay M, Ozbakkaloglu B. Increasing antimicrobial resistance in Escherichia coli isolates from community-acquired urinary tract infections during 1998-2003 in Manisa, Turkey. Japanese Journal of Infectious Diseases.
2005;
58
(3)
:
159-161
.
-
Astal ZE. Increasing ciprofloxacin resistance among prevalent urinary tract bacterial isolates in the Gaza Strip. Singapore Med J.
2005;
46
(9)
:
457-460
.
-
Kothari A, Sagar V. Antibiotic resistance in pathogens causing community-acquired urinary tract infections in India: a multicenter study. J Infect Dev Ctries.
2008;
2
(5)
:
354-358
.
View Article Google Scholar -
Ahmed AA, Osman H, Mansour AM, et al. Antimicrobial agent resistance in bacterial isolates from patients with diarrhea and urinary tract infection in the Sudan. The American Journal of Tropical Medicine and Hygiene.
2000;
63
(5)
:
259-263
.
View Article PubMed Google Scholar
Comments
Downloads
Article Details
Volume & Issue : Vol 7 No 8 (2020)
Page No.: 3944-3953
Published on: 2020-08-31
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 6040 times
- Download PDF downloaded - 1294 times
- View Article downloaded - 0 times
 Biomedpress
Biomedpress